Abstract
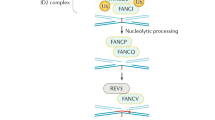
The Fanconi anaemia/Breast cancer consortium* Fanconi anaemia (FA) is an autosomal recessive disorder associated with progressive bone-marrow failure, a variety of congenital abnormalities, and predisposition to acute myeloid leukaemia1. Cells from FA patients show increased sensitivity to bifunctional DNA crosslinking agents such as diepoxybutane and mitomycin C, with characteristic chromosome breakage2. FA is genetically heterogeneous, at least five different complementation groups (FA-A to FA-E) having been described3,4. The gene for group C (FAC) was cloned by functional complementation and mapped to chromosome 9q22.3 (refs 3, 5), but the genes for the other complementation groups have not yet been identified. The group A gene (FAA) has recently been mapped to chromosome 16q24.3 by linkage analysis6, and accounts for 60–65% of FA cases7,8. We narrowed the candidate region by linkage and allelic association analysis, and have isolated a gene that is mutated in FA-A patients. The gene encodes a protein of 1,455 amino acids that has no significant homology to any other known proteins, and may therefore represent a new class of genes associated with the prevention or repair of DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Butturini, A. et al. Hematologic abnormalities in Fanconi anemia. An International Fanconi Anemia Registry Study. Blood 84, 1650–1655 (1994).
Auerbach, A.D. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp. Hematol. 21, 731–733 (1993).
Strathdee, C.A., Duncan, A.M.V. & Buchwald, M. Evidence for at least four Fanconi anaemia genes including FACC on chromosome9. Nature Genet. 1, 196–198 (1992).
Joenje, H. et al. Classification of Fanconi anaemia patients by complementation analysis: evidence for a fifth genetic subtype. Blood 86, 2156–2160 (1995).
Strathdee, C.A., Gavish, H., Shannon, W.R. & Buchwald, M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature 356, 763–767 (1992).
Pronk, J.C. et al. Localisation of the Fanconi anaemia complementation group A gene to chromosome 16q24.3. Nature Genet. 11, 338–340 (1995).
Buchwald, M. Complementation groups: one or more per gene? Nature Genet. 11, 228–239 (1995).
Gschwend, M. et al. A locus for Fanconi anemia on 16q determined by homozygosity of mapping. Am.J.Hum. Genet. 59, 377–384 (1996).
Doggett, N.A., Breuning, M.H. & Callen, D.F. Report of the fourth international workshop on human chromosome 16 mapping 1995. Cytogenet. Cell Genet. 72, 271–293 (1996).
Cleton-Jansen, A.M. et al. At least two different regions are involved in allelic imbalance on chromosome arm 16q in breast cancer. Genes, Chromosomes Cancer 9, 101–107 (1994).
Doggett, N.A. et al. An integrated physical map of human chromosome 16. Nature 377, 335–365 (1995).
Stallings, R.L. et al. Evaluation of a cosmid contig physical map of human chromosome 16. Genomics 13, 1031–1039 (1992).
Lennon, G., Auffray, C., Polymeropoulos, M. & Soares, M.B. The I.M.A.G.E. Consortium: an Integrated Molecular Analysis of Genomes and their Expression. Genomics 33, 151–152 (1996).
Savoia, A., Zatterale, A., Del Principe, D. & Joenje, H. Fanconi anaemia in Italy: high prevalence of complementation group A in two geographic clusters. Hum. Genet. 97, 599–603 (1996).
Lo Ten Foe, J.R. et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nature Genet. 14, 320–323 (1996).
Nakai, K., Kanehisa, M. A knowledge base for predicting protein localisation sites in eukaryotic cells. Genomics 14, 897–911 (1992).
Robbins, J., Dilworth, S.M., Laskey, R.A. & Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–23 (1991).
Trumpp, A., Blundell, P.A., de la Pompa, J.L. & Zeller, R. The chicken limb deformity gene encodes nuclear proteins expressed in specific cell types during morphogenesis. Genes Dev. 6, 14–28 (1992).
Verlander, P.C. et al. Mutation analysis of the Fanconi anemia gene FACC. Am. J. Hum. Genet. 54, 595–601 (1994).
Gibson, R.A. et al. Novel mutations and polymorphisms in the Fanconi anaemia group C gene. Hum. Mutat. 8, 140–148 (1996).
Liu, J.M., Buchwald, M., Walsh, C.E. & Young, N.S. Fanconi anemia and novel strategies for therapy. Blood 84, 3995–4007 (1994).
Dib, C. et al. The Généthon human genetic linkage map. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380, 152–154 (1996).
Lathrop, G.M., Lalouel, J.M., Julier, C. & Ott, J. Strategies for multilocus linkage analysis in humans. Proc. Natl. Acad. Sci. USA 81, 3443–3446 (1984).
Church, D.M. et al. Isolation of genes from complex sources of mammalian genomic DNA using exon amplification. Nature Genet. 6, 98–105 (1994).
Buckler, A.J. et al. Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc. Natl. Acad. Sci. USA 88, 4005–4009 (1991).
Tagle, D.A., Swaroop, M., Lovett, M. & Collins, F.S. Magnetic bead capture of expressed sequences encoded within large genomic segments. Nature 361, 751–753 (1993).
Sideras, P. et al. Transcription of unrearranged IgH chain genes in human B cell malignancies. Biased expression of genes encoded with the first duplication unit of the IgH chain locus. J. Immunol. 149, 244–252 (1992).
Gibson, R., Hajianpour, A., Murer-Orlando, M., Buchwald, M. & Mathew, C.G. A nonsense mutation and exon skipping in the Fanconi anaemia group C gene. Hum. Molec. Genet. 2, 797–799 (1993).
Orita, M., Sasaki, Y., Siya, T. & Hayashi, K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5, 874–879 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Apostolou, S., Whitmore, S., Crawford, J. et al. Positional cloning of the Fanconi anaemia group A gene. Nat Genet 14, 324–328 (1996). https://doi.org/10.1038/ng1196-324
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ng1196-324
This article is cited by
-
A case report and literature review of Fanconi Anemia (FA) diagnosed by genetic testing
Italian Journal of Pediatrics (2015)
-
Coregulation of FANCA and BRCA1 in human cells
SpringerPlus (2014)
-
Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins
Nature Reviews Genetics (2007)