Abstract
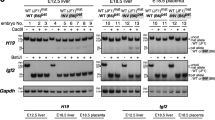
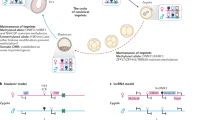
Genomic imprinting relies on establishing and maintaining the parental-specific methylation of DNA elements that control the differential expression of maternal and paternal alleles. Although the essential DNA methyltransferases have been discovered, proteins that regulate the sequence-specific establishment and maintenance of allelic methylation have not been identified. One candidate regulator of methylation, the zinc-finger protein CTCF, binds to the imprinting control region (ICR) of the genes Igf2 (encoding insulin-like growth factor 2) and H19 (fetal liver mRNA; refs. 1,2). The unmethylated maternal ICR is a chromatin boundary that prevents distant enhancers from activating Igf2 (refs. 3–6). In vitro experiments have suggested that CTCF mediates boundary activity of the maternal ICR, and that methylation of the paternal ICR abolishes this activity by preventing CTCF binding3,4,6. Using mice with point mutations in all four CTCF sites in the ICR, we show that maternally transmitted mutant ICRs in neonatal mice acquire a substantial but heterogeneous degree of methylation. Mutant ICRs in oocytes and blastocysts are not methylated, however, indicating that binding of CTCF is not required to establish the unmethylated ICR during oogenesis. We also show that the mutant ICR lacks enhancer-blocking activity, as the expression of Igf2 is activated on mutant maternal chromosomes. Conversely, maternal H19 expression is reduced, suggesting a positive role for CTCF in the transcription of that gene. This study constitutes the first in vivo demonstration of the multiple functions of CTCF in an ICR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Thorvaldson, J.L., Duran, K.L. & Bartolomei, M.S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12, 3693–3702 (1998).
Srivastava, M. et al. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis-acting regulatory region upstream of H19. Genes Dev. 14, 1186–1195 (2000).
Bell, A.C. & Felsenfeld, G. Modulation of an CTCF-dependent enhancer boundary by DNA methylation controls imprinting of the Igf2 gene. Nature 405, 482–485 (2000).
Hark, A.T. et al. CTCF mediates methylation-sensitive enhancer blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000).
Kaffer, C.R. et al. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14, 1908–1919 (2000).
Kanduri, C. et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853–856 (2000).
Bunting, M., Bernstein, K.E., Greer, J.M., Capecchi, M.R. & Thomas, K.R. Targeting genes for self-excision in the germ line. Genes Dev. 13, 1524–1528 (1999).
Hoess, R.H., Wierzbicki, A. & Abremski, K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 14, 2287–2300 (1986).
Olek, A. & Walter, J. The pre-implantation ontogeny of the H19 methylation imprint. Nat. Genet. 17, 275–276 (1997).
Tremblay, K.D., Saam, J.R., Ingram, R.S., Tilghman, S.M. & Bartolomei, M.S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9, 407–413 (1995).
Bourc'his, D., Xu, G.L., Lin, C.S., Bollman, B. & Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002).
Leighton, P.A., Ingram, R.S., Eggenschwiler, J., Efstratiadis, A. & Tilghman, S.M. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34–39 (1995).
Thorvaldsen, J.L., Mann, M.R., Nwoko, O., Duran, K.L. & Bartolomei, M.S. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22, 2450–2462 (2002).
Chao, W., Huynh, K.D., Spencer, R.J., Davidow, L.S. & Lee, J.T. CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295, 345–347 (2002).
Birger, Y., Shemer, R., Perk, J. & Razin, A. The imprinting box of the mouse Igf2r gene. Nature 397, 84–88 (1999).
Brandeis, M. et al. Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438 (1994).
Macleod, D., Charlton, J., Mullins, J. & Bird, A.P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8, 2282–2292 (1994).
Jirtle, R.L. Genomic imprinting and cancer. Exp. Cell Res. 248, 18–24 (1999).
Jones, B.K., Levorse, J. & Tilghman, S.M. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 12, 2200–2207 (1998).
Ramirez-Solis, R. et al. Genomic DNA microextraction: a method to screen numerous samples. Anal. Biochem. 201, 331–335 (1992).
Clark, S.J., Harrison, J., Paul, C.L. & Frommer, M. High-sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994).
Tremblay, K.D., Duran, K.L. & Bartolomei, M.S. A 5′ two kilobase pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17, 4322–4329 (1997).
Warnecke, P.M. et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 25, 4422–4426 (1997).
Olek, A., Oswald, J. & Walter, J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24, 5064–5066 (1996).
Acknowledgements
We thank R. Ingram for DNA sequencing; B. Jones for RNA samples; D. Mancini-DiNardo for advice on bisulphite analysis; M. Capecchi for the pACN plasmid; and members of S.M.T.'s laboratory for comments. This work was supported by a grant from the US National Institute for General Medical Sciences. S.M.T. was an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Schoenherr, C., Levorse, J. & Tilghman, S. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet 33, 66–69 (2003). https://doi.org/10.1038/ng1057
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1057
This article is cited by
-
The transgenic IG-DMR sequence of the mouse Dlk1-Dio3 domain acquired imprinted DNA methylation during the post-fertilization period
Epigenetics & Chromatin (2023)
-
CTCF modulates allele-specific sub-TAD organization and imprinted gene activity at the mouse Dlk1-Dio3 and Igf2-H19 domains
Genome Biology (2019)
-
Synthetic DNA fragments bearing ICR cis elements become differentially methylated and recapitulate genomic imprinting in transgenic mice
Epigenetics & Chromatin (2018)
-
YY1’s role in the Peg3 imprinted domain
Scientific Reports (2017)
-
Essential role of insulin-like growth factor 2 in resistance to histone deacetylase inhibitors
Oncogene (2016)