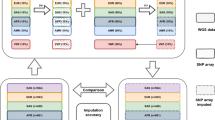
Abstract
Noncoding variants at human chromosome 9p21 near CDKN2A and CDKN2B are associated with type 2 diabetes1,2,3,4, myocardial infarction5,6,7, aneurysm8, vertical cup disc ratio9 and at least five cancers10,11,12,13,14,15,16. Here we compare approaches to more comprehensively assess genetic variation in the region. We carried out targeted sequencing at high coverage in 47 individuals and compared the results to pilot data from the 1000 Genomes Project. We imputed variants into type 2 diabetes and myocardial infarction cohorts directly from targeted sequencing, from a genotyped reference panel derived from sequencing and from 1000 Genomes Project low-coverage data. Polymorphisms with frequency >5% were captured well by all strategies. Imputation of intermediate-frequency polymorphisms required a higher density of tag SNPs in disease samples than is available on first-generation genome-wide association study (GWAS) arrays. Our association analyses identified more comprehensive sets of variants showing equivalent statistical association with type 2 diabetes or myocardial infarction, but did not identify stronger associations than the original GWAS signals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Saxena, R. et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 (2007).
Scott, L.J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–1345 (2007).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008).
Zeggini, E. et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316, 1336–1341 (2007).
Helgadottir, A. et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007).
Kathiresan, S. et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 (2009).
McPherson, R. et al. A common allele on chromosome 9 associated with coronary heart disease. Science 316, 1488–1491 (2007).
Helgadottir, A. et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat. Genet. 40, 217–224 (2008).
Ramdas, W.D. et al. A genome-wide association study of optic disc parameters. PLoS Genet. 6, e1000978 (2010).
Bishop, D.T. et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 41, 920–925 (2009).
Falchi, M. et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 41, 915–919 (2009).
Sherborne, A.L. et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat. Genet. 42, 492–494 (2010).
Shete, S. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 41, 899–904 (2009).
Stacey, S.N. et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 41, 909–914 (2009).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Wrensch, M. et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat. Genet. 41, 905–908 (2009).
Frazer, K.A. et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Li, Y. & Abecasis, G. MACH 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
Visel, A. et al. Targeted deletion of the 9p21 noncoding coronary artery disease risk interval in mice. Nature 464, 409–412 (2010).
Harismendy, O. et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470, 264–268 (2011).
Gnirke, A. et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 27, 182–189 (2009).
Stephens, M. & Scheet, P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76, 449–462 (2005).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Acknowledgements
Sample collections in the DGI study were funded by grants from the Sigrid Juselius and Folkhälsan foundations and from the Swedish Research Council (L.G.). The DGI GWAS study was supported by a grant from Novartis. The MIGen study was funded by the US National Institutes of Health (NIH) and the US National Heart, Lung, and Blood Institute's STAMPEED genomics research program through a grant to D.A. (R01 HL087676). S.K. is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award, a charitable gift from the Fannie E. Rippel Foundation, the Donovan Family Foundation, a career development award from the NIH and the Department of Medicine and Cardiovascular Research Center at Massachusetts General Hospital. D.A. and J.S. are supported in part by a Distinguished Clinical Scholar Award from the Doris Duke Charitable Foundation (to D.A.). Next-generation sequencing for this work was carried out by the Broad Institute Sequencing Platform, and genotyping was carried out by the Broad Institute Genetic Analysis Platform. We acknowledge their excellence and collaboration on this study. Sequencing was supported in part by a grant from the US National Human Genome Research Institute and by the Broad Institute. We thank M. Rivas, A. Sivachenco and K. Garimella for helpful discussions on sequencing, and B. Voight, S. Ripke and R. Do for helpful discussions on imputation.
Author information
Authors and Affiliations
Consortia
Contributions
J.S., V.A., A.A.P. and D.A. wrote the manuscript; L.G. and the Myocardial Infarction Genomics Consortium provided clinical samples; C.G., R.C.O., N.P.B. and S.G. contributed to next-generation sequencing data generation; A.A.P., J.M., E.B., M.D.P., S.G., M.J.D. and D.A. carried out sequencing analysis and variant calling; J.S., V.A., M.J.D. and D.A. carried out imputation and association analysis; J.S., V.A., B.T., C.G. and N.P.B. carried out genotyping and analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1, Supplementary Figures 1–11 and Supplementary Note. (PDF 5012 kb)
Supplementary Table 2
List of PCR primers and hybrid selection baits used in sequencing (XLS 268 kb)
Supplementary Table 3
List of all variants identified in high coverage sequencing (XLS 340 kb)
Supplementary Table 4
Validation analysis for SNPs identified in sequencing on 9p21 (XLS 112 kb)
Supplementary Table 5
List of variants in the genotyped reference panel for 9p21 (XLS 140 kb)
Supplementary Table 6
Imputation and association results for T2D and MI on 9p21 (XLS 275 kb)
Rights and permissions
About this article
Cite this article
Shea, J., Agarwala, V., Philippakis, A. et al. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat Genet 43, 801–805 (2011). https://doi.org/10.1038/ng.871
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.871
This article is cited by
-
The hazards of genotype imputation when mapping disease susceptibility variants
Genome Biology (2024)
-
The GEnetic Syntax Score: a genetic risk assessment implementation tool grading the complexity of coronary artery disease—rationale and design of the GESS study
BMC Cardiovascular Disorders (2021)
-
Dominant role of CDKN2B/p15INK4B of 9p21.3 tumor suppressor hub in inhibition of cell-cycle and glycolysis
Nature Communications (2021)
-
Re-analysis of public genetic data reveals a rare X-chromosomal variant associated with type 2 diabetes
Nature Communications (2018)
-
Islet biology, the CDKN2A/B locus and type 2 diabetes risk
Diabetologia (2016)