Abstract
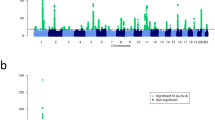
To identify new susceptibility loci for psoriasis, we undertook a genome-wide association study of 594,224 SNPs in 2,622 individuals with psoriasis and 5,667 controls. We identified associations at eight previously unreported genomic loci. Seven loci harbored genes with recognized immune functions (IL28RA, REL, IFIH1, ERAP1, TRAF3IP2, NFKBIA and TYK2). These associations were replicated in 9,079 European samples (six loci with a combined P < 5 × 10−8 and two loci with a combined P < 5 × 10−7). We also report compelling evidence for an interaction between the HLA-C and ERAP1 loci (combined P = 6.95 × 10−6). ERAP1 plays an important role in MHC class I peptide processing. ERAP1 variants only influenced psoriasis susceptibility in individuals carrying the HLA-C risk allele. Our findings implicate pathways that integrate epidermal barrier dysfunction with innate and adaptive immune dysregulation in psoriasis pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Nestle, F.O., Kaplan, D.H. & Barker, J. Psoriasis. N. Engl. J. Med. 361, 496–509 (2009).
Veal, C.D. et al. Family-based analysis using a dense single-nucleotide polymorphism-based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am. J. Hum. Genet. 71, 554–564 (2002).
Nair, R.P. et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am. J. Hum. Genet. 78, 827–851 (2006).
Capon, F. et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum. Mol. Genet. 17, 1938–1945 (2008).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
de Cid, R. et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 41, 211–215 (2009).
Hollox, E.J. et al. Psoriasis is associated with increased β-defensin genomic copy number. Nat. Genet. 40, 23–25 (2008).
Nair, R.P. et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 41, 199–204 (2009).
Zhang, X.J. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999).
Jenisch, S. et al. Linkage analysis of human leukocyte antigen (HLA) markers in familial psoriasis: strong disequilibrium effects provide evidence for a major determinant in the HLA-B/-C region. Am. J. Hum. Genet. 63, 191–199 (1998).
York, I.A. et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat. Immunol. 3, 1177–1184 (2002).
Burton, P.R. et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Khan, M.A. Update on spondyloarthropathies. Ann. Intern. Med. 136, 896–907 (2002).
Qian, Y. et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8, 247–256 (2007).
Hayden, M.S. & Ghosh, S. Shared principles in NF-kappaB signaling. Cell 132, 344–362 (2008).
Gregersen, P.K. et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 41, 820–823 (2009).
Oyamada, A. et al. Tyrosine kinase 2 plays critical roles in the pathogenic CD4 T cell responses for the development of experimental autoimmune encephalomyelitis. J. Immunol. 183, 7539–7546 (2009).
Wallace, C. et al. The imprinted DLK1–MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 42, 68–71 (2010).
Zhou, Z. et al. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J. Virol. 81, 7749–7758 (2007).
Kotenko, S.V. et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77 (2003).
Wilkins, C. & Gale, M. Jr. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22, 41–47 (2010).
Nejentsev, S., Walker, N., Riches, D., Egholm, M. & Todd, J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 (2009).
Lincoln, M.R. et al. Epistasis among HLA-DRB1, HLA-DQA1, and HLA-DQB1 loci determines multiple sclerosis susceptibility. Proc. Natl. Acad. Sci. USA 106, 7542–7547 (2009).
Orsmark-Pietras, C. et al. Biological and genetic interaction between tenascin C and neuropeptide S receptor 1 in allergic diseases. Hum. Mol. Genet. 17, 1673–1682 (2008).
Smyth, D.J. et al. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes 57, 1730–1737 (2008).
The Wellcome Trust Case-Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007).
Picard, C. et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. Eur. J. Immunol. 39, 1966–1976 (2009).
Leslie, S., Donnelly, P. & McVean, G. A statistical method for predicting classical HLA alleles from SNP data. Am. J. Hum. Genet. 82, 48–56 (2008).
Barrett, J.C. et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41, 1330–1334 (2009).
R Development Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2010).
Schouten, E.G. et al. Risk ratio and rate ratio estimation in case-cohort designs: hypertension and cardiovascular mortality. Stat. Med. 12, 1733–1745 (1993).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Su, Z. et al. A Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association studies. Stat. Sci. 24, 430–450 (2009).
Marchini, J., Donnelly, P. & Cardon, L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 37, 413–417 (2005).
de Bakker, P.I. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Stephens, M., Smith, N.J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Acknowledgements
The principal funding for this study was provided by the Wellcome Trust, as part of the Wellcome Trust Case Control Consortium 2 project (083948/Z/07/Z). We also thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, A. Wilk, C.R. Stribling and S. Taylor of the Wellcome Trust Sanger Institute's Sample and Genotyping Facilities for technical assistance. We thank D. Davison for making available his program 'Shellfish' for calculating principal components in large genetic datasets. Case collections were supported by the Netherlands Organization for Health Research and Development (P.L.J.M.Z.); the Swedish Medical Research Council, Karolinska Institutet, Karolinska University Hospital, Psoriasis Foundation, AFA Insurance and Welander Finsen Foundation (M.S.); the Association for the Defence of Psoriasis Patients (G.N.); Psoriasis Association and the Cecil King Memorial Foundation (M.J.C.); the Swedish Psoriasis Association (L.S.); the German Research Foundation (Tr 228/5-4 and Re 679/10-4) and The Interdisciplinary Centre for Clinical Research (IZKF B32/A8) of the University of Erlangen-Nuremberg (A. Reis); the Spanish Ministry of Science and Innovation (grant SAF 2008-00357) and the 'Generalitat de Catalunya' Departments of Health and Universities and Innovation (X.E.); the Genetic Repository in Ireland for Psoriasis and Psoriatic Arthritis (GRIPPsA), the Dublin Centre for Clinical Research (DCCR, funded by the Irish Health Research Board), The Wellcome Trust and Science Foundation Ireland (R. McManus); National Institute for Health Research, Manchester Biomedical Research Centre (J.W., C.E.M.G., R.B.W., H.S.Y.); and Arthritis Research UK (J.W., grant 17552). P. Donnelly was supported in part by a Wolfson-Royal Society Merit Award, and A.O. was supported by a PhD studentship from The Generation Trust. We also acknowledge support from the UK Medical Research Council (to R.C.T., F.O.N., A.H. and J.N.B., grant G0601387), the Wellcome Trust (F.O.N., grant 078173/Z/05/Z) and the Department of Health through the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre awards to Guy's and St. Thomas' National Health Service (NHS) Foundation Trust in partnership with King's College London (J. Knight., M.E.W., C.G.M., F.O.N., A. Hayday and J.N.B.) and the NIHR award to Moorfields Eye Hospital NHS Foundation Trust and University College London Institute of Ophthalmology for a Specialist Biomedical Research Centre for Ophthalmology (A.C.V.). We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02, and of the UK National Blood Service controls funded by the Wellcome Trust. We thank W. Bodmer and B. Winney for use of the People of the British Isles DNA collection, which was funded by the Wellcome Trust.
Author information
Authors and Affiliations
Consortia
Contributions
F.C., A.D.B., C.E.M.G., J. Kere, A. Reis, J.N.B. and R.C.T. oversaw cohort collection for both the discovery and the replication datasets. The WTCCC2 DNA, genotyping, data quality control and informatics group (S.J.B., P. Deloukas, S.E., E. Gray, S.E.H., C.L. and S.C.P.) executed GWAS sample handling, genotyping and quality control. Members of the WTCCC2 analysis group (C.C.A.S., A.S., G.B., C.B., C.F., M.P., Z.S. and P. Donnelly), J. Knight and M.E.W. performed statistical analyses. A. Dilthey, S.L., L. Moutsianas and G.M. performed HLA imputation and analyses. D.M.E. and M.A.B. provided advice on similarities of association to another autoimmune disease. A.S., F.C., C.C.A.S., J. Knight, M.E.W., A. Hayday, J.N.B., P. Donnelly and R.C.T. contributed to writing the manuscript. The WTCCC2 Management Committee (P. Donnelly (chair), J.M.B., E.B., J.P.C., A. Duncanson, J.J., H.S.M., C.G.M., C.N.A.P., R.P., S.J.S., A. Rautanen, A.C.V., N.W., M.A.B., L.P. and R.C.T.) monitored the execution of the GWAS. The GAP Consortium (R.C.T. (chair), M.H.A., A.B., J.G.M.B., M.J.C., A.C., X.E., O.F., E. Giardina, A. Hofer, U.H., A.D.I., B.K., J. Lascorz, J. Leman, L. Mallbris, W.H.I.M., R. McManus, R. Mössner, Å.T.N., F.O.N., G.N., A.O., C.P., R.M.P., E.R.M., A.W.R., W.S., L.S., J.S., C.H.S., M.S., R.T.A., H.T., R.B.W., W.W., K.W., J.W., H.S.Y. and P.L.J.M.Z.) contributed to sample collection. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–4 and Supplementary Note. (PDF 6704 kb)
Supplementary Table 9
Other SNPs with GWAS p-value less than 10-4 (XLS 37 kb)
Rights and permissions
About this article
Cite this article
Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42, 985–990 (2010). https://doi.org/10.1038/ng.694
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.694
This article is cited by
-
Reassessing human MHC-I genetic diversity in T cell studies
Scientific Reports (2024)
-
Non-mutational neoantigens in disease
Nature Immunology (2024)
-
Immunosuppression causes dynamic changes in expression QTLs in psoriatic skin
Nature Communications (2023)
-
Disentangling the complexity of psoriasis in the post-genome-wide association era
Genes & Immunity (2023)
-
Air pollution exposure and auto-inflammatory and autoimmune diseases of the musculoskeletal system: a review of epidemiologic and mechanistic evidence
Environmental Geochemistry and Health (2023)