Abstract
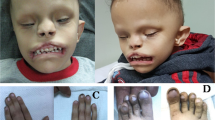
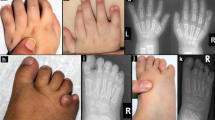
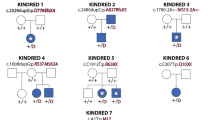
Arhinia, or absence of the nose, is a rare malformation of unknown etiology that is often accompanied by ocular and reproductive defects. Sequencing of 40 people with arhinia revealed that 84% of probands harbor a missense mutation localized to a constrained region of SMCHD1 encompassing the ATPase domain. SMCHD1 mutations cause facioscapulohumeral muscular dystrophy type 2 (FSHD2) via a trans-acting loss-of-function epigenetic mechanism. We discovered shared mutations and comparable DNA hypomethylation patterning between these distinct disorders. CRISPR/Cas9-mediated alteration of smchd1 in zebrafish yielded arhinia-relevant phenotypes. Transcriptome and protein analyses in arhinia probands and controls showed no differences in SMCHD1 mRNA or protein abundance but revealed regulatory changes in genes and pathways associated with craniofacial patterning. Mutations in SMCHD1 thus contribute to distinct phenotypic spectra, from craniofacial malformation and reproductive disorders to muscular dystrophy, which we speculate to be consistent with oligogenic mechanisms resulting in pleiotropic outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
20 March 2017
In the version of this article initially published, the legend to Figure 4c stated that only one proband without SMCHD1 mutation was tested for D4Z4 methylation pattern. However, three probands and one affected family member without SMCHD1 mutation were tested, as shown in the figure. The error has been corrected in the HTML and PDF versions of the article.
References
Bosma, J.F., Henkin, R.I., Christiansen, R.L. & Herdt, J.R. Hypoplasia of the nose and eyes, hyposmia, hypogeusia, and hypogonadotrophic hypogonadism in two males. J. Craniofac. Genet. Dev. Biol. 1, 153–184 (1981).
Hogan, B.L. et al. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J. Embryol. Exp. Morphol. 97, 95–110 (1986).
Glaser, T. et al. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 7, 463–471 (1994).
Schmidt-Sidor, B. et al. Malformations of the brain in two fetuses with a compound heterozygosity for two PAX6 mutations. Folia Neuropathol. 47, 372–382 (2009).
Solomon, B.D. et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am. J. Med. Genet. A. 149A, 2543–2546 (2009).
Gordon, C.T. et al. De novo mutations in SMCHD1 cause Bosma arhinia microphthalmia syndrome and abrogate nasal development. Nat. Genet. http://dx.doi.org/10.1038/ng.3765 (2016).
Parhar, I.S. Cell migration and evolutionary significance of GnRH subtypes. Prog. Brain Res. 141, 3–17 (2002).
Cho, C.-H., Shakibaei, M., Merker, H.-J. & Klein, M. The rare malformation of nasal aplasia. Mund Kiefer Gesichtschir. 10, 106–117 (2006).
Thiele, H., Musil, A., Nagel, F. & Majewski, F. Familial arhinia, choanal atresia, and microphthalmia. Am. J. Med. Genet. 63, 310–313 (1996).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Samocha, K.E. et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 46, 944–950 (2014).
Blewitt, M.E. et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 40, 663–669 (2008).
Chen, K. et al. Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation. Proc. Natl. Acad. Sci. USA 112, E3535–E3544 (2015).
Gendrel, A.V. et al. Epigenetic functions of Smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 33, 3150–3165 (2013).
Mould, A.W. et al. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin 6, 19 (2013).
Lemmers, R.J. et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 44, 1370–1374 (2012).
Hirano, T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7, 311–322 (2006).
Lemmers, R.J. et al. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum. Mol. Genet. 24, 659–669 (2015).
van den Boogaard, M.L. et al. Double SMCHD1 variants in FSHD2: the synergistic effect of two SMCHD1 variants on D4Z4 hypomethylation and disease penetrance in FSHD2. Eur. J. Hum. Genet. 24, 78–85 (2016).
Lemmers, R.J. et al. Hemizygosity for SMCHD1 in facioscapulohumeral muscular dystrophy type 2: consequences for 18p deletion syndrome. Hum. Mutat. 36, 679–683 (2015).
Chen, K. et al. The epigenetic regulator Smchd1 contains a functional GHKL-type ATPase domain. Biochem. J. 473, 1733–1744 (2016).
Jones, T.I. et al. Identifying diagnostic DNA methylation profiles for facioscapulohumeral muscular dystrophy in blood and saliva using bisulfite sequencing. Clin. Epigenetics 6, 23 (2014).
Jones, T.I. et al. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin. Epigenetics 7, 37 (2015).
Liu, C. et al. A secreted WNT-ligand-binding domain of FZD5 generated by a frameshift mutation causes autosomal dominant coloboma. Hum. Mol. Genet. 25, 1382–1391 (2016).
Chassaing, N. et al. Targeted resequencing identifies PTCH1 as a major contributor to ocular developmental anomalies and extends the SOX2 regulatory network. Genome Res. 26, 474–485 (2016).
Yahyavi, M. et al. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum. Mol. Genet. 22, 3250–3258 (2013).
Steven, C. et al. Molecular characterization of the GnRH system in zebrafish (Danio rerio): cloning of chicken GnRH-II, adult brain expression patterns and pituitary content of salmon GnRH and chicken GnRH-II. Gen. Comp. Endocrinol. 133, 27–37 (2003).
Whitlock, K.E., Illing, N., Brideau, N.J., Smith, K.M. & Twomey, S. Development of GnRH cells: setting the stage for puberty. Mol. Cell. Endocrinol. 254-255, 39–50 (2006).
Zohar, Y., Muñoz-Cueto, J.A., Elizur, A. & Kah, O. Neuroendocrinology of reproduction in teleost fish. Gen. Comp. Endocrinol. 165, 438–455 (2010).
Abraham, E., Palevitch, O., Gothilf, Y. & Zohar, Y. The zebrafish as a model system for forebrain GnRH neuronal development. Gen. Comp. Endocrinol. 164, 151–160 (2009).
Sharpe, J. et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science 296, 541–545 (2002).
Blewitt, M.E. et al. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl. Acad. Sci. USA 102, 7629–7634 (2005).
Kelley, L.A., Mezulis, S., Yates, C.M., Wass, M.N. & Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Chen, J., Bardes, E.E., Aronow, B.J. & Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009).
Hall, J.G. Pena–Shokeir phenotype (fetal akinesia deformation sequence) revisited. Birth Defects Res. A Clin. Mol. Teratol. 85, 677–694 (2009).
Solomon, B.D., Gropman, A. & Muenke, M. Holoprosencephaly overview. GeneReviews https://www.ncbi.nlm.nih.gov/books/NBK1530/ (updated 29 August 2013).
Lederer, D. et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 90, 119–124 (2012).
Lindgren, A.M. et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum. Genet. 132, 537–552 (2013).
Lahiry, P. et al. A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am. J. Hum. Genet. 84, 134–147 (2009).
de Greef, J.C. et al. Clinical features of facioscapulohumeral muscular dystrophy 2. Neurology 75, 1548–1554 (2010).
van Deutekom, J.C. et al. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum. Mol. Genet. 5, 1997–2003 (1996).
Calandra, P. et al. Allele-specific DNA hypomethylation characterises FSHD1 and FSHD2. J. Med. Genet. 53, 348–355 (2016).
van den Boogaard, M.L. et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am. J. Hum. Genet. 98, 1020–1029 (2016).
Weemaes, C.M. et al. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur. J. Hum. Genet. 21, 1219–1225 (2013).
Albers, C.A. et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 44, 435–439 (2012).
Timberlake, A.T. et al. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. eLife 5, e20125 (2016).
Fokkema, I.F. et al. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 32, 557–563 (2011).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Petersen, B., Petersen, T.N., Andersen, P., Nielsen, M. & Lundegaard, C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9, 51 (2009).
Yang, J. et al. The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Heffernan, R. et al. Improving prediction of secondary structure, local backbone angles, and solvent accessible surface area of proteins by iterative deep learning. Sci. Rep. 5, 11476 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Van der Auwera, G.A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
O'Leary, N.A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Turner, S.D. qqman: an R package for visualizing GWAS results using Q–Q and Manhattan plots. Preprint at. bioRxiv http://dx.doi.org/10.1101/005165 (2014).
Blumenthal, I. et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am. J. Hum. Genet. 94, 870–883 (2014).
Sugathan, A. et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA 111, E4468–E4477 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Wu, T.D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
DeLuca, D.S. et al. RNA-SeQC: RNA–seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA–seq experiments. Bioinformatics 28, 2184–2185 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Aken, B.L. et al. The Ensembl gene annotation system. Database (Oxford) 2016, baw093 (2016).
Fay, M.P. & Shaw, P.A. Exact and asymptotic weighted logrank tests for interval censored data: the interval R package. J. Stat. Softw. 36, i02 (2010).
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016).
Eppig, J.T., Blake, J.A., Bult, C.J., Kadin, J.A. & Richardson, J.E. The Mouse Genome Database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 43, D726–D736 (2015).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA–seq data with DESeq2. Genome Biol. 15, 550 (2014).
Rohde, C., Zhang, Y., Reinhardt, R. & Jeltsch, A. BISMA—fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics 11, 230 (2010).
Lemmers, R.J. et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010).
Lemmers, R.J. et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 81, 884–894 (2007).
Lemmers, R.J. et al. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am. J. Hum. Genet. 86, 364–377 (2010).
Lemmers, R.J., O'Shea, S., Padberg, G.W., Lunt, P.W. & van der Maarel, S.M. Best practice guidelines on genetic diagnostics of facioscapulohumeral muscular dystrophy: workshop 9th June 2010, LUMC, Leiden, the Netherlands. Neuromuscul. Disord. 22, 463–470 (2012).
Kague, E. et al. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS One 7, e47394 (2012).
Niederriter, A.R. et al. In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp. 78, e50338 (2013).
Labun, K., Montague, T.G., Gagnon, J.A., Thyme, S.B. & Valen, E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276 (2016).
Isrie, M. et al. Mutations in either TUBB or MAPRE2 cause circumferential skin creases Kunze type. Am. J. Hum. Genet. 97, 790–800 (2015).
Acknowledgements
We thank all participants, family members and clinical staff for their generous contributions of time and materials to this research. We thank T. Gillis, J. Ruliera, C. Hanscom, C. Antolik and M. Anderson for technical assistance. This project was funded by grants from the National Institutes of Health ((NIH) R00MH095867 and R01HD081256 to M.E.T.; P01GM061354 to M.E.T., J.F.G., C.C.M. and E.C.L.; T32HD007396 to H. Brand; P50HD028138 to W.F.C., S.B.S., M.E.T., N.K. and E.E.D.; R01HD043341 and MGH Robert and Laura Reynolds Research Scholar Award to S.B.S.; K23HD073304-02 and 1SI2ES025429-01 to N.D.S.; P50DK096415 to N.K. and R01AR062587 to P.L.J.); the March of Dimes (FY15-255 to M.E.T.); the Medical Research Council (MR/M02122X/1 to J.A.M.); the German Research Foundation (SFB665 to A.M.K.) and the Berlin Institute of Health (BIH-CRG1 to A.M.K.). D.R.F., R.R.M. (MC_PC_U127574433), D.S.D., H. Bengani, K.A.W., J.R., J.K.R. and J.A.M. are funded by program grants from the Medical Research Council (MRC) Human Genetics Unit award to the University of Edinburgh. M.A. is funded by the University of Edinburgh Institute of Genomics and Molecular Medicine Translational Initiative Fund. S.A.M. is supported by U54-NS053672, which funds the Iowa, Paul D. Wellstone Muscular Dystrophy Cooperative Research Center. N.K. is supported as a Distinguished Jean and George Brumley Professor at Duke University, and M.E.T. is supported as the Desmond and Ann Heathwood MGH Research Scholar.
Author information
Authors and Affiliations
Contributions
M.E.T., D.R.F., E.E.D., N.K., P.J., N.D.S. and H. Brand designed the study. N.D.S., L.P., K.A.W., M.N., S.P., T.K., D.L., A. Silva, S.J., J.C.S., M.F.L., S.S.S., N.P., J.R.L., N.F., A.V., A.R., K. Steindl, I.S., D.S., N.O., C.J., J.T., S.C., L.A.S., B.B., C. Cesaretti, J.E.G.-O., T.P.B., O.P.S., J.D.H., W.M., K.W.R., B.L.L., M.S., A.M.K., C.-H.C., C.C.M., V.v.H., R.B., J.E.H., S.B.S., K.Y., J.M.G., A.E.L., W.F.C. and D.R.F. recruited patients and collected clinical information and samples. Z.A.K., H. Bengani, L.P., S.E., T.I.J., J.R.W., J.R., A. Stortchevoi, C.M.S., Y.A., B.B.C., M.A., R.R.M., J.K.R., M.Z., J.W.J., E.C.L., S.A.M., N.K., P.L.J., E.E.D., D.R.F. and D.S.D. performed molecular genetics and animal modeling studies. H. Brand, K. Samocha, R.L.C., C. Chiang, A.L., M.L., J.F.G., D.G.M. and M.E.T. performed genomic analyses. J.A.M. performed protein modeling. N.D.S., H. Brand, N.K., J.F.G., P.L.J., E.E.D., D.R.F. and M.E.T. wrote the manuscript, which was revised and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 (PDF 5639 kb)
Supplementary Table 1
Arhina cohort-wide phenotype information. (XLSX 16 kb)
Supplementary Table 2
Individual level phenotype information. (XLSX 22 kb)
Supplementary Table 3
Ethnic specific analysis of SMCHD1 mutation frequency. (XLSX 9 kb)
Supplementary Table 4
DNA methylation analysis of D4Z4 repeats in arhinia cases and familial controls. (XLSX 17 kb)
Supplementary Table 5
Representative zebrafish morphometric data. (XLSX 21 kb)
Supplementary Table 6
Mouse CRISPR of SMCHD1 missense mutations. (XLSX 11 kb)
Supplementary Table 7
Allele-specific expression of SMCHD1 in subjects with a mutation. (XLSX 10 kb)
Supplementary Table 8
Differential expression analysis between arhinia cases and familial controls. (XLSX 1601 kb)
Supplementary Table 9
Enrichment of nominal human genes (P < 0.05) against mouse Smchd1 targets and differentially expressed genes in Smchd1-null mouse. (XLSX 10 kb)
Supplementary Table 10
Overlapping genes with significant dysregulation in the same direction in mouse and human. (XLSX 10 kb)
Supplementary Table 11
Primers used in study. (XLSX 11 kb)
Rights and permissions
About this article
Cite this article
Shaw, N., Brand, H., Kupchinsky, Z. et al. SMCHD1 mutations associated with a rare muscular dystrophy can also cause isolated arhinia and Bosma arhinia microphthalmia syndrome. Nat Genet 49, 238–248 (2017). https://doi.org/10.1038/ng.3743
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3743
This article is cited by
-
Non-coding autoimmune risk variant defines role for ICOS in T peripheral helper cell development
Nature Communications (2024)
-
Cholesterol biosynthesis modulates differentiation in murine cranial neural crest cells
Scientific Reports (2023)
-
SMCHD1 and LRIF1 converge at the FSHD-associated D4Z4 repeat and LRIF1 promoter yet display different modes of action
Communications Biology (2023)
-
Facioscapulohumeral muscular dystrophy: the road to targeted therapies
Nature Reviews Neurology (2023)
-
SMCHD1 has separable roles in chromatin architecture and gene silencing that could be targeted in disease
Nature Communications (2023)