Abstract
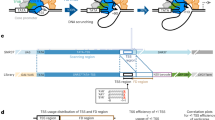
Mammalian transcriptomes are complex and formed by extensive promoter activity. In addition, gene promoters are largely divergent and initiate transcription of reverse-oriented promoter upstream transcripts (PROMPTs). Although PROMPTs are commonly terminated early, influenced by polyadenylation sites, promoters often cluster so that the divergent activity of one might impact another. Here we found that the distance between promoters strongly correlates with the expression, stability and length of their associated PROMPTs. Adjacent promoters driving divergent mRNA transcription support PROMPT formation, but owing to polyadenylation site constraints, these transcripts tend to spread into the neighboring mRNA on the same strand. This mechanism to derive new alternative mRNA transcription start sites (TSSs) is also evident at closely spaced promoters supporting convergent mRNA transcription. We suggest that basic building blocks of divergently transcribed core promoter pairs, in combination with the wealth of TSSs in mammalian genomes, provide a framework with which evolution shapes transcriptomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Kapranov, P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007).
Taft, R.J. et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 41, 572–578 (2009).
Preker, P. et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322, 1851–1854 (2008).
Core, L.J., Waterfall, J.J. & Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008).
Seila, A.C. et al. Divergent transcription from active promoters. Science 322, 1849–1851 (2008).
Sigova, A.A. et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. USA 110, 2876–2881 (2013).
Ntini, E. et al. Polyadenylation site–induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 20, 923–928 (2013).
Almada, A.E., Wu, X., Kriz, A.J., Burge, C.B. & Sharp, P.A. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363 (2013).
Core, L.J. et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat. Genet. 46, 1311–1320 (2014).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Nojima, T. et al. Mammalian NET-Seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161, 526–540 (2015).
Scruggs, B.S. et al. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 58, 1101–1112 (2015).
Flynn, R.A., Almada, A.E., Zamudio, J.R. & Sharp, P.A. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc. Natl. Acad. Sci. USA 108, 10460–10465 (2011).
Kaida, D. et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2010).
Duttke, S.H. et al. Human promoters are intrinsically directional. Mol. Cell 57, 674–684 (2015).
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Andersson, R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014).
Andersson, R. et al. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat Commun. 5, 5336 (2014).
Trinklein, N.D. et al. An abundance of bidirectional promoters in the human genome. Genome Res. 14, 62–66 (2004).
Li, Y.-Y. et al. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput. Biol. 2, e74 (2006).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005).
Engström, P.G. et al. Complex loci in human and mouse genomes. PLoS Genet. 2, e47 (2006).
Lehner, B., Williams, G., Campbell, R.D. & Sanderson, C.M. Antisense transcripts in the human genome. Trends Genet. 18, 63–65 (2002).
Andersson, R. et al. Human gene promoters are intrinsically bidirectional. Mol. Cell 60, 346–347 (2015).
Kadonaga, J.T. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol. 1, 40–51 (2012).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Thurman, R.E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Fenouil, R. et al. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 22, 2399–2408 (2012).
Pugh, B.F. & Venters, B.J. Genomic organization of human transcription initiation complexes. PLoS One 11, e0149339 (2016).
Rhee, H.S. & Pugh, B.F. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012).
Valen, E. et al. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat. Struct. Mol. Biol. 18, 1075–1082 (2011).
Pelechano, V., Wei, W. & Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 497, 127–131 (2013).
Lubas, M. et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011).
Faghihi, M.A. & Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 10, 637–643 (2009).
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005).
Wu, X. & Sharp, P.A. Divergent transcription: a driving force for new gene origination? Cell 155, 990–996 (2013).
Jensen, T.H., Jacquier, A. & Libri, D. Dealing with pervasive transcription. Mol. Cell 52, 473–484 (2013).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
FANTOM Consortium and the RIKEN PMI and CLST (DGT). A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014).
Karolchik, D. et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42, D764–D770 (2014).
Andersen, P.R. et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 20, 1367–1376 (2013).
Pelechano, V., Wei, W., Jakob, P. & Steinmetz, L.M. Genome-wide identification of transcript start and end sites by transcript isoform sequencing. Nat. Protoc. 9, 1740–1759 (2014).
Anders, S., Pyl, P.T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Wu, T.D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Marstrand, T.T. et al. Asap: a framework for over-representation statistics for transcription factor binding sites. PLoS ONE 3, e1623 (2008).
Frith, M.C. et al. A code for transcription initiation in mammalian genomes. Genome Res. 18, 1–12 (2008).
Carninci, P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635 (2006).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Robinson, J.T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Acknowledgements
Work in the T.H.J. laboratory was supported by the European Research Council (ERC) (grant 339953) as well as the Danish National Research Foundation (grant DNRF58), the Lundbeck Foundation and the Novo Nordisk Foundation. Work in the A.S. laboratory was supported by grants from the Lundbeck Foundation, the Novo Nordisk-Foundation and the Innovation Fund Denmark. J.H. was supported by a Boehringer-Ingelheim PhD fellowship. R.A. was supported by ERC grant 638273. Work in the L.M.S. laboratory was supported by the Deutsche Forschungsgemeinschaft (grant 1422/3-1). A.A.P. was supported by a Jane Coffin Childs postdoctoral fellowship. We thank the European Molecular Biology Laboratory Genomics Core Facility for technical support.
Author information
Authors and Affiliations
Contributions
Y.C. analyzed the data. A.A.P. performed exploratory computational analyses for divergent mRNAs. M.L. performed exploratory computational analyses for convergent loci constellations. N.M. and V.P. produced the TIF-seq libraries. A.I.J. processed TIF-seq reads. J.H. conducted RT-qPCR validations. R.A. assisted with enhancer definitions, co-guidance of analyses and interpretation of results. L.M.S. supervised A.I.J. and V.P.. Y.C. and A.S. produced images. T.H.J. and A.S. conceived and supervised the project. Y.C., T.H.J. and A.S. wrote the paper. All authors read and approved of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–2 and Supplementary Note 1 (PDF 20121 kb)
Supplementary Dataset 1
Genomic coordinates for analyzed regions. (XLSX 388 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Pai, A., Herudek, J. et al. Principles for RNA metabolism and alternative transcription initiation within closely spaced promoters. Nat Genet 48, 984–994 (2016). https://doi.org/10.1038/ng.3616
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3616
This article is cited by
-
Bidirectional promoters: an enigmatic genome architecture and their roles in cancers
Molecular Biology Reports (2021)
-
Deep learning suggests that gene expression is encoded in all parts of a co-evolving interacting gene regulatory structure
Nature Communications (2020)
-
A Novel Divergent Gene Transcription Paradigm—the Decisive, Brain-Specific, Neural |-Srgap2–Fam72a-| Master Gene Paradigm
Molecular Neurobiology (2019)
-
Engineered bidirectional promoters enable rapid multi-gene co-expression optimization
Nature Communications (2018)
-
ChIP-seq and ChIP-exo profiling of Pol II, H2A.Z, and H3K4me3 in human K562 cells
Scientific Data (2018)