Abstract
Dun is a wild-type coat color in horses characterized by pigment dilution with a striking pattern of dark areas termed primitive markings. Here we show that pigment dilution in Dun horses is due to radially asymmetric deposition of pigment in the growing hair caused by localized expression of the T-box 3 (TBX3) transcription factor in hair follicles, which in turn determines the distribution of hair follicle melanocytes. Most domestic horses are non-dun, a more intensely pigmented phenotype caused by regulatory mutations impairing TBX3 expression in the hair follicle, resulting in a more circumferential distribution of melanocytes and pigment granules in individual hairs. We identified two different alleles (non-dun1 and non-dun2) causing non-dun color. non-dun2 is a recently derived allele, whereas the Dun and non-dun1 alleles are found in ancient horse DNA, demonstrating that this polymorphism predates horse domestication. These findings uncover a new developmental role for T-box genes and new aspects of hair follicle biology and pigmentation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
NCBI Reference Sequence
Referenced accessions
NCBI Reference Sequence
References
Wade, C.M. et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867 (2009).
Orlando, L. et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013).
Bowling, A.T. & Ryder, O.A. Genetic studies of blood markers in Przewalski's horses. J. Hered. 78, 75–80 (1987).
Adalsteinsson, S. Inheritance of yellow dun and blue dun in the Icelandic toelter horse. J. Hered. 69, 146–148 (1978).
Bamshad, M. et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat. Genet. 16, 311–315 (1997).
Frank, D.U., Emechebe, U., Thomas, K.R. & Moon, A.M. Mouse TBX3 mutants suggest novel molecular mechanisms for Ulnar-mammary syndrome. PLoS One 8, e67841 (2013).
Kumar, P.P. et al. TBX3 regulates splicing in vivo: a novel molecular mechanism for Ulnar-mammary syndrome. PLoS Genet. 10, e1004247 (2014).
Gremmel, F. Coat colors in horses. J. Hered. 30, 437–445 (1939).
Andersson, L.S. et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646 (2012).
Schubert, M. et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci. USA 111, E5661–E5669 (2014).
Thomas-Chollier, M. et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 6, 1860–1869 (2011).
Ma, L. et al. 'Cyclic alopecia' in Msx2 mutants: defects in hair cycling and hair shaft differentiation. Development 130, 379–389 (2003).
Kayserili, H. et al. ALX4 dysfunction disrupts craniofacial and epidermal development. Hum. Mol. Genet. 18, 4357–4366 (2009).
Reginelli, A.D., Wang, Y.Q., Sassoon, D. & Muneoka, K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development 121, 1065–1076 (1995).
Satokata, I. et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24, 391–395 (2000).
Steingrímsson, E., Copeland, N.G. & Jenkins, N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38, 365–411 (2004).
Kunisada, T. et al. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development 125, 2915–2923 (1998).
Yoshida, H. et al. Review: melanocyte migration and survival controlled by SCF/c-kit expression. J. Investig. Dermatol. Symp. Proc. 6, 1–5 (2001).
St-Jacques, B. et al. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8, 1058–1068 (1998).
van den Boogaard, M. et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Invest. 122, 2519–2530 (2012).
Fang, M., Larson, G., Ribeiro, H.S., Li, N. & Andersson, L. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341 (2009).
Pruvost, M. et al. Genotypes of predomestic horses match phenotypes painted in Paleolithic works of cave art. Proc. Natl. Acad. Sci. USA 108, 18626–18630 (2011).
Gabriel, S.B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Sundström, E. et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genomics 13, 365 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Schubert, M. et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082 (2014).
Lindgreen, S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337 (2012).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Delaneau, O., Marchini, J. & Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Acknowledgements
We thank the numerous horse owners who provided samples, T. Raudsepp and I. Randlaht for samples from Estonian native horses, C. Asa and M. Fischer (St. Louis Zoo) and F. Marshall for providing hair samples from the Somali wild ass, W. Zimmerman for photographs of Przewalski's horse, S. Fard, H. Ring and F. Hallböök for advice on histological characterization, O. Ryder, L. Chemnick and C. Steiner for delivering DNA extracts from Przewalski's horses, C. Der Sarkissian and L. Ermini for assistance in whole-genome resequencing at the Centre for GeoGenetics, Denmark, and the HudsonAlpha Genomic Services Laboratory for RNA-seq. This work was supported by grants from the Knut and Alice Wallenberg foundation (to L.A.) and the US National Institutes of Health (to G.S.B.), as well as by an Erasmus Mundus fellowship within the framework of the European Graduate School of Animal Breeding and Genetics (to D.S.). Sequencing was performed by the SNP&SEQ Technology Platform, supported by Uppsala University and Hospital, the Science for Life Laboratory and the Swedish Research Council (80576801 and 70374401).
Author information
Authors and Affiliations
Contributions
L.A. led the genetic characterization and G.S.B. led the RNA-seq and immunohistochemistry studies. F.I., P.I., K.M. and M.C.T.P. did the sampling. F.I. was responsible for phenotyping and carried out genotyping together with U.G., L.M. and M.C.T.P. K.M. performed immunohistochemistry analysis. C.H. performed RNA-seq analysis. C.-J.R., F.I., J.B., M.S. and L.O. were responsible for genome sequence analysis. E.S. analyzed transcription factor binding sites. D.S. contributed to TBX3 expression analysis. F.I., L.A., M.C.T.P., K.L.-T., G.L., S.M. and C.W. took part in the initial mapping of Dun. L.A., F.I., K.M., C.H. and G.S.B. wrote the manuscript with input from other authors. All authors approved the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Dun phenotypes in domestic horses.
(a) Dun versus non-dun phenotype on eumelanic, eumelanic and phaeomelanic, and phaeomelanic backgrounds. Genotypes at the Dun (D, d), Extension (E, e) and Agouti (A, a) loci are indicated. In clockwise order from the upper left: blue Dun, bay Dun, red Dun, chestnut, bay and black. (b) Various primitive markings in Duns. (c) Red Dun (D/- e/e) as camouflage. (d) Non-dun chestnut (d/d e/e) failing as camouflage. (Photographs by Freyja Imsland and Páll Imsland.)
Supplementary Figure 2 non-dun1 and non-dun2 phenotypes in domestic horses.
(a) Three horses with different genotypes at the Dun locus on black base color (E/- a/a) and transverse sections through hairs from the dorsal stripe/midline and croup of each horse. Colored arrows indicate the sampling locations of hairs and punch biopsies: yellow, dorsal midline; red, croup. Scale bar, 50 μm. (b) Bay (E/- A/-) homozygous non-dun1/non-dun1 horse, shown from two different angles on the same day. (c) Bay (E/- A/-) heterozygous non-dun1/non-dun2 horse, showing faint dorsal stripe, pronounced leg barring and a faint shadow of a shoulder stripe. (Photographs by Freyja Imsland and Páll Imsland.)
Supplementary Figure 3 TBX3, TBX5 and KITLG mRNA expression in skin from Dun and non-dun horses.
(a) Relative mRNA levels (mean ± s.e.m.) for TBX3 and TBX5 as assessed by RT-qPCR from Dun/- (n = 7), non-dun1/non-dun1 (n = 3) and non-dun2/non-dun2 (n = 6) croup skin. (b) Relative mRNA levels (mean ± s.e.m.) for KITLG mRNA from Dun/- (n = 7) and non-dun/non-dun (n = 9; 2 d1/d1 and 7 d2/d2 samples) croup skin. The results of two tailed t tests are indicated in the figure.
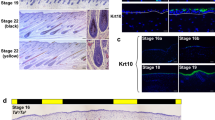
Supplementary Figure 4 TBX3 immunofluorescence in Dun and non-dun hair follicles.
(a) Five-micrometer serial sections from Dun/- croup skin. (b) Sections from dorsal midline skin of Dun/-, non-dun1/non-dun1 and non-dun2/non-dun2 horses. TBX3 immunofluorescence is shown in green, pigment is pseudocolored red in b, and white lines mark the basement membrane. Scale bars, 50 μm.
Supplementary Figure 5 MITF and KITLG immunofluorescence in Dun and non-dun skin.
(a) Immunofluorescence for MITF (green) on croup skin sections from Dun/-, non-dun1/non-dun1 and non-dun2/non-dun2 horses. Top panels show the pigmented epidermis (red, pseudocolor), and the white line marks the dermal (D)-epidermal (E) junction. Corresponding bright-field photomicrographs are on the right in the lower panel, White and black lines mark the basement membrane. (b) Immunofluorescence staining for KITLG (green) on Dun/- and non-dun2/non-dun2 croup skin. Bright-field images showing epidermal pigment are on the right. The white line marks the dermal-epidermal junction. DAPI staining is in blue. Scale bars, 50 μm (a) and 100 μm (b).
Supplementary Figure 6 TBX3 immunolocalization relative to markers of hair follicle differentiation.
(a) Anatomy of the hair bulb (left; Me, medulla; Co, cortex/hair cuticle; IRS, inner root sheath; Cp, companion layer; ORS, outer root sheath; Pro, proliferative cells; Dp, dermal papilla). Cellular compartments are marked by the hair follicle differentiation markers AE15, AE13 and K6 (KRT6, keratin 6). Ki-67 marks proliferative cells. Immunofluorescence for AE15 (green; middle) and K6 (green; right) on anagen hair follicles from non-dun (d2/d2) horses. (b) Immunofluorescence for TBX3 (green; left) and AE13 (red; middle) from Dun (D/-) horses. The merged image is on the right. Pigment is pseudocolored (red), and DAPI staining is blue in a. White lines mark the basement membrane in b. Scale bar in b, 100 μm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1, 2, 4 and 7. (PDF 2069 kb)
Supplementary Table 3: Summary of results of SNP screen using 384 SNPs from the TBX3 region.
Genotypes of the SNPs most strongly associated with the dun and non-dun phenotypes, marked by red dots in Figure 2e. The results of three different association tests for dominant genotypes are reported: (i) Dun versus non-dun1 including both domestic and Przewalski's horses, (ii) Dun versus non-dun1 including only domestic horses and (iii) Dun versus non-dun (non-dun1 and non-dun2 combined) including both domestic and Przewalski's horses. All but four SNPs could be excluded as being causative on the basis of the genotypes of Przewalski's horses. Two of those four could also be excluded on the basis of the presence of a recombinant Dun haplotype in domestic horse 7. The genotype was confirmed via Sanger sequencing in this individual and in two related dun horses that also proved recombinant. "nc" indicates no call. (XLSX 62 kb)
Supplementary Table 5: Haplotype analysis of the 40-kb region centered at the non-dun2 deletion on the basis of the SNP screen.
Individuals shown are representative for the data set. "nc" indicates no call. (XLSX 65 kb)
Supplementary Table 6: Detailed results of the RNA-seq differential gene expression study in skin from dun and non-dun horses.
(a) Samples obtained from the croup. (b) Samples obtained from the dorsal midline. Four columns are included for each detected gene, displaying the average overall expression level (normalized gene counts) and a log2-transformed fold change between dun and non-dun samples, as well as the degree of statistical significance of the observed differences expressed as P values and q values (adjusted for multiple testing using the Benjamini-Hochberg correction approach). Genes are sorted according to the observed P values in the analysis of croup samples. (XLSX 2198 kb)
Rights and permissions
About this article
Cite this article
Imsland, F., McGowan, K., Rubin, CJ. et al. Regulatory mutations in TBX3 disrupt asymmetric hair pigmentation that underlies Dun camouflage color in horses. Nat Genet 48, 152–158 (2016). https://doi.org/10.1038/ng.3475
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3475
This article is cited by
-
Transcriptomic analyses during development reveal mechanisms of integument structuring and color production
Evolutionary Ecology (2023)
-
Characteristic of Przewalski horses population from Askania-Nova reserve based on genetic markers
Molecular Biology Reports (2023)
-
Hair follicle regional specificity in different parts of bay Mongolian horse by histology and transcriptional profiling
BMC Genomics (2020)
-
A novel sex-linked mutant affecting tail formation in Hongshan chicken
Scientific Reports (2017)
-
Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array
Scientific Reports (2017)