Abstract
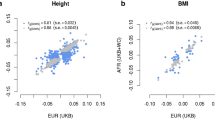
Across-nation differences in the mean values for complex traits are common1,2,3,4,5,6,7,8, but the reasons for these differences are unknown. Here we find that many independent loci contribute to population genetic differences in height and body mass index (BMI) in 9,416 individuals across 14 European countries. Using discovery data on over 250,000 individuals and unbiased effect size estimates from 17,500 sibling pairs, we estimate that 24% (95% credible interval (CI) = 9%, 41%) and 8% (95% CI = 4%, 16%) of the captured additive genetic variance for height and BMI, respectively, reflect population genetic differences. Population genetic divergence differed significantly from that in a null model (height, P < 3.94 × 10−8; BMI, P < 5.95 × 10−4), and we find an among-population genetic correlation for tall and slender individuals (r = −0.80, 95% CI = −0.95, −0.60), consistent with correlated selection for both phenotypes. Observed differences in height among populations reflected the predicted genetic means (r = 0.51; P < 0.001), but environmental differences across Europe masked genetic differentiation for BMI (P < 0.58).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Moussavi, S. et al. Depression, chronic diseases, and decrements in health: results from the World Health Surveys. Lancet 370, 851–858 (2007).
Wild, S., Roglic, G., Green, A., Sicree, R. & King, H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27, 1047–1053 (2004).
Dye, C. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. J. Am. Med. Assoc. 282, 677–686 (1999).
Lopez, A.D., Mathers, C.D., Ezzati, M., Jamison, D.T. & Murray, C.J.L. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 367, 1747–1757 (2006).
Wang, H. et al. Age-specific and sex-specific mortality in 187 countries, 1970–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2071–2094 (2012).
Jemal, A., Center, M.M., DeSantis, C. & Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomarkers Prev. 19, 1893–1907 (2010).
Kim, A.S. & Johnston, S.C. Global variation in the relative burden of stroke and ischemic heart disease. Circulation 124, 314–323 (2011).
Johnston, S.C., Mendis, S. & Mathers, C.D. Global variation in stroke burden and mortality: estimates from monitoring, surveillance, and modelling. Lancet Neurol. 8, 345–354 (2009).
Yang, J., Visscher, P.M. & Wray, N.R. Sporadic cases are the norm for complex disease. Eur. J. Hum. Genet. 18, 1039–1043 (2010).
Hill, W.G., Goddard, M.E. & Visscher, P.M. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4, e1000008 (2008).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Morris, A.P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Lee, S.H. et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum. Mol. Genet. 22, 832–841 (2013).
Yang, J. et al. Ubiquitous polygenicity of human complex traits: genome-wide analysis of 49 traits in Koreans. PLoS Genet. 9, e1003355 (2013).
Robinson, M.R., Wray, N.R. & Visscher, P.M. Explaining additional genetic variation in complex traits. Trends Genet. 30, 124–132 (2014).
Abegunde, D.O., Mathers, C.D., Adam, T., Ortegon, M. & Strong, K. The burden and costs of chronic diseases in low-income and middle-income countries. Lancet 370, 1929–1938 (2007).
Kim, A.S. & Johnston, S.C. Temporal and geographic trends in the global stroke epidemic. Stroke 44, S123–S125 (2013).
Ezzati, M. & Riboli, E. Can noncommunicable diseases be prevented? Lessons from studies of populations and individuals. Science 337, 1482–1487 (2012).
Hartl, D.L. & Clark, A.G. Principles of Population Genetics (Sinauer Associates, 1997).
Leinonen, T., McCairns, R.J.S., O'Hara, R.B. & Merilä, J. QST-FST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat. Rev. Genet. 14, 179–190 (2013).
James, P.T., Rigby, N. & Leach, R. The obesity epidemic, metabolic syndrome and future prevention strategies. Eur. J. Cardiovasc. Prev. Rehabil. 11, 3–8 (2004).
Popkin, B.M. Global nutrition dynamics: the world is shifting rapidly toward a diet linked with noncommunicable diseases. Am. J. Clin. Nutr. 84, 289–298 (2006).
Wang, Y.C., McPherson, K., Marsh, T., Gortmaker, S.L. & Brown, M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 378, 815–825 (2011).
Ng, M. et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 384, 766–781 (2014).
Finucane, M.M. et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9·1 million participants. Lancet 377, 557–567 (2011).
Turchin, M.C. et al. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat. Genet. 44, 1015–1019 (2012).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Yang, J. et al. FTO genotype is associated with phenotypic variability of body mass index. Nature 490, 267–272 (2012).
Amato, R., Miele, G., Monticelli, A. & Cocozza, S. Signs of selective pressure on genetic variants affecting human height. PLoS ONE 6, e27588 (2011).
Berg, J.J. & Coop, G. A population genetic signal of polygenic adaptation. PLoS Genet. 10, e1004412 (2014).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Locke, A.E. et al. Genetic studies of body mass index yeild new insights for obesity biology. Nature 518, 197–206 (2015).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Dudbridge, F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013).
Baten, J. & Blum, M. Growing tall but unequal: new findings and new background evidence on anthropometric welfare in 156 countries, 1810–1989. Econ. Hist. Dev. Reg. 27, S66–S85 (2012).
Sabeti, P.C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007).
Nielsen, R., Hellmann, I., Hubisz, M., Bustamante, C. & Clark, A.G. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 8, 857–868 (2007).
Bustamante, C.D. et al. Natural selection on protein-coding genes in the human genome. Nature 437, 1153–1157 (2005).
Blekhman, R. et al. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18, 883–889 (2008).
Barreiro, L.B., Laval, G., Quach, H., Patin, E. & Quintana-Murci, L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40, 340–345 (2008).
Akey, J.M. et al. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2, e286 (2004).
Barreiro, L.B. & Quintana-Murci, L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11, 17–30 (2010).
Vasseur, E. & Quintana-Murci, L. The impact of natural selection on health and disease: uses of the population genetics approach in humans. Evol. Appl. 6, 596–607 (2013).
Chiaroni, J., Underhill, P.A. & Cavalli-Sforza, L.L. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc. Natl. Acad. Sci. USA 106, 20174–20179 (2009).
Ovaskainen, O., Karhunen, M., Zheng, C., Arias, J.M.C. & Merilä, J. A new method to uncover signatures of divergent and stabilizing selection in quantitative traits. Genetics 189, 621–632 (2011).
Diverse Populations Collaborative Group. Weight-height relationships and body mass index: some observations from the Diverse Populations Collaboration. Am. J. Phys. Anthropol. 128, 220–229 (2005).
Lande, R. Genetic variation and phenotypic evolution during allopatric speciation. Am. Nat. 116, 463–479 (1980).
Esko, T. et al. Genetic characterization of northeastern Italian population isolates in the context of broader European genetic diversity. Eur. J. Hum. Genet. 21, 659–665 (2013).
Weir, B.S. & Hill, W.G. Estimating F-statistics. Annu. Rev. Genet. 36, 721–750 (2002).
Weir, B.S. & Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Cockerham, C.C. & Weir, B.S. Correlations, descent measures: drift with migration and mutation. Proc. Natl. Acad. Sci. USA 84, 8512–8514 (1987).
Williams, A.L., Patterson, N., Glessner, J., Hakonarson, H. & Reich, D. Phasing of many thousands of genotyped samples. Am. J. Hum. Genet. 91, 238–251 (2012).
Howie, B., Marchini, J. & Stephens, M. Genotype imputation with thousands of genomes. G3 (Bethesda) 1, 457–470 (2011).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Delaneau, O., Marchini, J. & Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2012).
Hadfield, J.D. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33(2), 1–22 (2010)
Acknowledgements
We thank the reviewers for their very helpful and insightful comments that greatly improved the manuscript. We also thank B. Hill and O. Ovaskainen for useful discussions. The University of Queensland group is supported by the Australian National Health and Medical Research Council (NHMRC; grants 1078037, 1048853 and 1050218). J.E.P. is supported by Australian Research Council grant DE130100691. J.Y. is supported by a Charles and Sylvia Viertel Senior Medical Research Fellowship and by NHMRC grant 1052684. We thank our colleagues at the Centre for Neurogenetics and Statistical Genomics for comments and suggestions. We are grateful to the twins and their families for their generous participation in the full-sibling family data set, which includes data from many cohorts and received support from many funding bodies. TWINGENE was supported by the Swedish Research Council (M-2005-1112), GenomEUtwin (EU/QLRT-2001-01254 and QLG2-CT-2002-01254), US National Institutes of Health (NIH) grant DK U01-066134, the Swedish Foundation for Strategic Research (SSF), and the Heart and Lung Foundation (20070481). For the Netherlands Twin Register (NTR), funding was obtained from the Netherlands Organization for Scientific Research (NWO; MagW/ZonMW grants 904-61-090, 985-10-002, 904-61-193, 480-04-004, 400-05-717, Addiction-31160008, Middelgroot-11-09-032 and Spinozapremie 56-464-14192), the Center for Medical Systems Biology (CSMB; NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL; 184.021.007), the VU University's Institute for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA), the European Science Foundation (ESF; EU/QLRT-2001-01254), the European Community's Seventh Framework Programme (FP7/2007-2013) under the ENGAGE project grant agreement (HEALTH-F4-2007-201413), the European Research Council (ERC Advanced; 230374), the Rutgers University Cell and DNA Repository (National Institute for Mental Health (NIMH), U24-MH068457-06), the Avera Institute (Sioux Falls, South Dakota, USA) and the US NIH (R01-D0042157-01A, Grand Opportunity grants 1RC2-MH089951-01 and 1RC2-MH089995-01). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. The TwinsUK study was funded by the Wellcome Trust and the European Community's Seventh Framework Programme (FP7/2007-2013) under the ENGAGE project grant agreement (HEALTH-F4-2007-201413). TwinsUK also receives support from the UK Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's and St Thomas' National Health Service (NHS) Foundation Trust in partnership with King's College London. T.D.S. is the holder of an ERC Advanced Principal Investigator award. Genotyping for the TwinsUK study was performed by the Wellcome Trust Sanger Institute, with the support of the National Eye Institute via a US NIH/Center for Inherited Disease Research (CIDR) genotyping project. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (contract N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI contract N02-HL-64278. Funding for SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. The QIMR researchers acknowledge funding from the Australian NHMRC (grants 241944, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 496688 and 552485) and the US NIH (grants AA07535, AA10248, AA014041, AA13320, AA13321, AA13326 and DA12854). We are grateful to M. Gill (Trinity College Dublin) and K. Nicodemus (University of Edinburgh) for access to the ISC–Trinity College Dublin cohort, which was supported by the Wellcome Trust and the Health Research Board, Ireland. Access to the Bulgarian cohort data was kindly facilitated by G. Kirov and V. Excott-Price. For the Danish cohort, the Danish Scientific Committees and the Danish Data Protection Agency approved the study and all the patients gave written informed consent before inclusion in the project. The National Institute on Aging (NIA) provided funding for the Health and Retirement Study (HRS; U01-AG09740). The HRS is performed at the Institute for Social Research at the University of Michigan. This manuscript was not prepared in collaboration with investigators of the HRS and does not necessarily reflect the opinions or views of the HRS, University of Michigan or NIA. The Netherlands genotype samples were part of Project MinE, which was supported by the ALS Foundation Netherlands. Research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013).
Author information
Authors and Affiliations
Contributions
Conception and design of the study: M.R.R., M.E.G., J.Y. and P.M.V. Data analysis: M.R.R., with additional contributions from G.H., C.M.-G., M.M., K.S., T.E., J.E.P., A.V., S.I.B., S.G., A.E.J., B.K., A.E.L., T.H.P., S.V., A.R.W. and W.v.R. Study oversight, sample collection and management: J.H.V., L.H.v.d.B., O.A.A., P.G., A.M., F.R., T.M.W., G.R.A., D.I.B., D.I.C., E.J.C.d.G., T.M.F., J.N.H., J.J.H., E.I., R.J.F.L., P.K.E.M., N.G.M., G.W.M., K.E.N., N.L.P., T.D.S. and E.K.S. Manuscript writing: M.R.R. and P.M.V., with contributions from all authors on the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 2 Projection of the prediction samples onto the first two HapMap principal components.
For the non-ascertained set of independent (LD r2 < 0.1), common, HapMap 3 loci that we used for prediction, we projected our prediction samples onto the first two principal components from HapMap.
Supplementary Figure 3 Simulation study comparing population and within-family association testing.
Using real genotype data, causal variants were allocated to 5,000 independent loci at random across the genome, with effects sampled from a normal distribution. Effect sizes were estimated in a genome-wide association study without controlling for population stratification (GWAS; yellow), in a GWAS that controlled for the first 20 principal components (green) and in a sibling pair within-family design (blue). The effect sizes were then used to predict the phenotype in an independent set of sibling pair data, and a recently derived approach was used to test for population stratification bias in the effect size estimates (Online Methods and ref. 33). Variance attributable to a Cg or a Ce term is indicative of population stratification bias, which can be observed in the GWAS scenario that did not appropriately control for population stratification.
Supplementary Figure 4 Simulation study comparing population and within-family association.
Using real genotype data, causal variants were allocated to 5,000 independent loci at random across the genome, with their effects sampled from a normal distribution. Fifty simulation replicates were conducted. Effect sizes were estimated in a genome-wide association study without controlling for population stratification (GWAS) and in a sibling pair within-family design. The effect sizes from the top 100 independent SNPs identified by the GWAS, the top 500 independent SNPs identified by the GWAS and genome-wide independent loci were then used for prediction in an independent sample. Individuals from the prediction sample were projected onto the first principal component of the discovery sample, and then two groups were selected based on the upper and lower quartiles of the distribution of the projected principal component. The mean difference in the predictor is shown when the predictor was created using effect sizes estimated in a GWAS that did not control for population stratification, using effect sizes estimated in a within-family design, using the true simulated effect sizes and when the within-family effect sizes were randomly allocated to loci under our null model. A predictor created from within-family estimates of effect size yields similar estimates to the true simulated values and the null model, and in no simulation were our predictions significantly different from our null model. This result was irrespective of whether there was ascertainment of loci from a discovery GWAS containing population stratification bias, as when we selected the top 100 or top 500 loci from the GWAS, re-estimated the effects in a within-family analysis and created a predictor we found no evidence of differentiation from our null model. A predictor created from GWAS estimates of effect size that are biased by population stratification yields variable estimates of the prediction difference between the 2 groups, and in 10 of the 50 simulations our predictions differed significantly from our null model.
Supplementary Figure 5 Simulation study comparing population and within-family association testing showing potential ascertainment bias.
Using real genotype data, causal variants were allocated to 5,000 independent loci at random across the genome, with their effects sampled from a normal distribution. Fifty simulation replicates were conducted. Genotype-environment correlation was induced through a phenotypic mean difference of 0.5 s.d. along the first principal component of the discovery sample. Effect sizes were estimated in a genome-wide association study without controlling for population stratification (GWAS) and in a sibling pair within-family design. The effect sizes from the top 100 independent SNPs identified by the GWAS, the top 500 independent SNPs identified by the GWAS and genome-wide independent loci were then used for prediction in an independent sample. Individuals from the prediction sample were projected onto the first principal component of the discovery sample, and two groups were then selected based on the upper and lower quartiles of the distribution of the projected principal component. The mean difference in the predictor is shown when the predictor was created using effect sizes estimated in a GWAS that did not control for population stratification, using effect sizes estimated in a within-family design, using the true simulated effect sizes and when the within-family effect sizes were randomly allocated to loci under our null model. Ascertainment bias is evident here, where if the top 100 or 500 loci from a GWAS containing population stratification were selected to create a predictor then, irrespective of whether biased SNP effect estimates from the GWAS or unbiased SNP effect estimates from the within-family analysis were used, a significant deviation from the null model is observed. This is because the loci selected from the GWAS were those where the genotype-environment correlation was the strongest. In this simulation scenario, there is no selection on the phenotype and, thus, when there is no ascertainment of loci (when genome-wide SNPs are used to create the predictor), no prediction differences from the true values or the null model were evident.
Supplementary Figure 6 Proportion of population-level variance in a genetic predictor comprised of different sets of SNPs for height and BMI.
Genetic predictors were created from independent (pairwise LD correlation < 0.1, >1 Mb apart), common HapMap 3 loci selected at different significance thresholds (P < 5 × 10–8, P < 5 × 10–6, P < 5 × 10–4, P < 0.005, P < 0.05, P < 0.1) from large-scale meta-analyses. All refers to genome-wide independent (pairwise LD correlation < 0.1, >1 Mb apart), common HapMap 3 loci that were either selected preferentially on the basis of their within-family association with either trait or selected at random.
Supplementary Figure 7 Increased prediction accuracy results in increased population-level variance.
We selected SNPs from large-scale meta-analyses, re-estimated their effects in a within-family design that is unbiased of population stratification and then assessed prediction accuracy in an independent sample for (a) height and (b) body mass index. We find that the population-level variance is better captured when a greater amount of phenotypic variation is explained by the predictor.
Supplementary Figure 9 The characteristics of a SNP and its contribution to the genome-wide pattern of population differentiation for height.
Relationship between the contribution of a SNP to the genome-wide pattern of population differentiation (c2 value) and (a) allele frequency, (b) phenotypic variance explained and (c) meta-analysis P value.
Supplementary Figure 10 The characteristics of a SNP and its contribution to the genome-wide pattern of population differentiation for body mass index.
Relationship between the contribution of a SNP to the genome-wide pattern of population differentiation (c2 value) and (a) allele frequency, (b) phenotypic variance explained and (c) meta-analysis P value.
Supplementary Figure 11 Simulation study of the general approach.
(a) Simulated distribution of allele frequency differentiation, θ, at 10,000 loci plotted against allele frequency. (b) The simulated association between θ and the additive genetic variance contributed by each locus, which assumes that the most differentiated loci are those that contribute most to the additive genetic variance. (c) Profile scores were calculated from different sets of simulated loci, where each set explained differing amounts of the total variance. Population-level variance is shown for each set of loci, demonstrating that, even when the loci cumulatively explain only 20% of the total variance, 50% of the population-level genetic variance is captured. (d) Error variance for each locus was simulated from a normal distribution with variance equal to a percentage of the total variance in profile score. Increasing amounts of error variance were added to the profile score to approximate the effects of adding an increasing number of false positive loci. Population-level variance is shown at different error variances, demonstrating that including a large number of false positives decreases the population-level effects.
Supplementary Figure 12 Simulation study of the error variance induced by the addition of null SNPs.
The error variance for each locus was simulated from a normal distribution as a percentage of the total variance in profile score. Increasing amounts of error variance were added to the profile score to approximate the effects of adding an increasing number of false positive loci. The 95% confidence intervals are shown for the population mean profile score of 11 simulated populations across increasing amounts of incorporated error variance: (a) 0%, (b) 1%, (c) 2.5%, (d) 5%, (e) 7.5% and (f) 10%. This pattern reflects the fact that including a large number of false positives decreases the amount of population-level variance estimated.
Supplementary Figure 13 Overlap in the annotation of differentiated loci to genes for height and BMI.
Five hundred SNP loci were selected that are expected to contribute most to the pattern of population genetic differentiation for height and body mass index. These SNP loci were annotated to genes, and the overlap was estimated. The annotation was then repeated by randomly selecting 500 loci from the top 10,000 SNP loci 100 times. The 95% confidence interval of the percentage of overlapping genes across the 100 sampling steps was 8.4–19.4%. In the expected top 500 loci contributing to population genetic variation of each trait, the percentage overlap was 19.8%.
Supplementary Figure 14 Population genetic differentiation across six Italian villages.
Predicted population genetic differentiation for BMI and height on a small scale across six northern Italian villages. There was no significant differentiation from the null model for height (c2 = 0.23, P = 0.985) and for BMI (c2 = 6.27, P = 0.817) at any set of SNPs, and we present the results across the genome. The villages are Sauris (sa), Resia (re), Illegio (il), Erto (er), Clausetto (cl) and San Martino del Corso (sm) in the Friuli-Venezia Giulia region in northeastern Italy. On this small scale, we find no evidence for population differentiation for either phenotype.
Supplementary Figure 15 Population genetic differentiation in the Human Genetic Diversity Panel for independent genome-wide SNPs ascertained on within-family effect sizes.
(a) Height and (b) BMI. P values give the deviation of the predicted means from the null expectation. The proportion of population-level variance was 17.5% (95% CI = 9.6, 27.9) for height and 13.1% (95% CI = 7.2, 21.3) for BMI. Worldwide, there was no evidence of any population-level genetic correlation (0.007, 95% CI = –0.051, 0.064).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1 and 2, and Supplementary Note. (PDF 6893 kb)
Rights and permissions
About this article
Cite this article
Robinson, M., Hemani, G., Medina-Gomez, C. et al. Population genetic differentiation of height and body mass index across Europe. Nat Genet 47, 1357–1362 (2015). https://doi.org/10.1038/ng.3401
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3401
This article is cited by
-
Assessment of the Long-Term Exposure to Lead in Four European Countries Using PBPK Modeling
Exposure and Health (2024)
-
Changes of anthropometric indicators of lithuanian first-graders in 2008–2019 according to International Obesity Task Force (IOTF) and World Health Organization (WHO) definitions
BMC Public Health (2023)
-
Detecting associated genes for complex traits shared across East Asian and European populations under the framework of composite null hypothesis testing
Journal of Translational Medicine (2022)
-
Genomic approaches to trace the history of human brain evolution with an emerging opportunity for transposon profiling of ancient humans
Mobile DNA (2021)
-
Evaluating marginal genetic correlation of associated loci for complex diseases and traits between European and East Asian populations
Human Genetics (2021)