Abstract
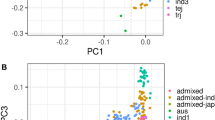
Plant metabolites are important to world food security in terms of maintaining sustainable yield and providing food with enriched phytonutrients. Here we report comprehensive profiling of 840 metabolites and a further metabolic genome-wide association study based on ∼6.4 million SNPs obtained from 529 diverse accessions of Oryza sativa. We identified hundreds of common variants influencing numerous secondary metabolites with large effects at high resolution. We observed substantial heterogeneity in the natural variation of metabolites and their underlying genetic architectures among different subspecies of rice. Data mining identified 36 candidate genes modulating levels of metabolites that are of potential physiological and nutritional importance. As a proof of concept, we functionally identified or annotated five candidate genes influencing metabolic traits. Our study provides insights into the genetic and biochemical bases of rice metabolome variation and can be used as a powerful complementary tool to classical phenotypic trait mapping for rice improvement.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Referenced accessions
European Nucleotide Archive
Sequence Read Archive
References
Saito, K. & Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489 (2010).
Schwab, W. Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62, 837–849 (2003).
Keurentjes, J.J. Genetical metabolomics: closing in on phenotypes. Curr. Opin. Plant Biol. 12, 223–230 (2009).
De Luca, V., Salim, V., Atsumi, S.M. & Yu, F. Mining the biodiversity of plants: a revolution in the making. Science 336, 1658–1661 (2012).
Hellmann, H. & Mooney, S. Vitamin B6: a molecule for human health? Molecules 15, 442–459 (2010).
Herrero, S., Gonzalez, E., Gillikin, J.W., Velez, H. & Daub, M.E. Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol. Biol. 76, 157–169 (2011).
Kaur, H., Heinzel, N., Schottner, M., Baldwin, I.T. & Galis, I. R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol. 152, 1731–1747 (2010).
Luo, J. et al. A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21, 318–333 (2009).
Butelli, E. et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 (2008).
Niggeweg, R., Michael, A.J. & Martin, C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746–754 (2004).
Luo, J. et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326 (2008).
Morohashi, K. et al. A genome-wide regulatory framework identifies maize Pericarp Color1 controlled genes. Plant Cell 24, 2745–2764 (2012).
Keurentjes, J.J. et al. The genetics of plant metabolism. Nat. Genet. 38, 842–849 (2006).
Huang, X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39 (2012).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010).
Gong, L. et al. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 110, 20320–20325 (2013).
Yamamoto, T., Yonemaru, J. & Yano, M. Towards the understanding of complex traits in rice: substantially or superficially? DNA Res. 16, 141–154 (2009).
Matsuda, F. et al. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 152, 566–578 (2010).
Riedelsheimer, C. et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 109, 8872–8877 (2012).
Chan, E.K., Rowe, H.C., Corwin, J.A., Joseph, B. & Kliebenstein, D.J. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9, e1001125 (2011).
Chan, E.K., Rowe, H.C., Hansen, B.G. & Kliebenstein, D.J. The complex genetic architecture of the metabolome. PLoS Genet. 6, e1001198 (2010).
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800 (2005).
Caicedo, A.L. et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3, 1745–1756 (2007).
Han, B. & Xue, Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr. Opin. Plant Biol. 6, 134–138 (2003).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Heuberger, A.L. et al. Metabolomic and functional genomic analyses reveal varietal differences in bioactive compounds of cooked rice. PLoS ONE 5, e12915 (2010).
Kawaura, K. et al. Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genomics 10, 271 (2009).
Kusano, M. et al. Deciphering starch quality of rice kernels using metabolite profiling and pedigree network analysis. Mol. Plant 5, 442–451 (2012).
Redestig, H. et al. Exploring molecular backgrounds of quality traits in rice by predictive models based on high-coverage metabolomics. BMC Syst. Biol. 5, 176 (2011).
Weng, J.K., Li, Y., Mo, H. & Chapple, C. Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337, 960–964 (2012).
Lippert, C. et al. FaST linear mixed models for genome-wide association studies. Nat. Methods 8, 833–835 (2011).
Atwell, S. et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010).
Chan, E.K., Rowe, H.C. & Kliebenstein, D.J. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185, 991–1007 (2010).
Shimizu, T. et al. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 287, 19315–19325 (2012).
Swaminathan, S., Morrone, D., Wang, Q., Fulton, D.B. & Peters, R.J. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21, 3315–3325 (2009).
Ko, J.H. et al. Four glucosyltransferases from rice: cDNA cloning, expression, and characterization. J. Plant Physiol. 165, 435–444 (2008).
Saitoh, K., Onishi, K., Mikami, I., Thidar, K. & Sano, Y. Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168, 997–1007 (2004).
Cheng, A.X. et al. The rice (E)-β-caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry 68, 1632–1641 (2007).
Evers, D. et al. Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J. Exp. Bot. 61, 2327–2343 (2010).
Minorsky, P.V. The hot and the classic. Plant Physiol. 128, 1167–1168 (2002).
Lehmann, T. & Pollmann, S. Gene expression and characterization of a stress-induced tyrosine decarboxylase from Arabidopsis thaliana. FEBS Lett. 583, 1895–1900 (2009).
Marinova, K. et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19, 2023–2038 (2007).
Johns, M.A. & Mao, L. Differentiation of the two rice subspecies indica and japonica: a Gene Ontology perspective. Funct. Integr. Genomics 7, 135–151 (2007).
Jung, K.H. et al. Genome-wide identification and analysis of japonica and indica cultivar-preferred transcripts in rice using 983 Affymetrix array data. Rice 6, 19 (2013).
Brachi, B., Morris, G.P. & Borevitz, J.O. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12, 232 (2011).
Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 (2011).
Zhao, K. et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2, 467 (2011).
Morris, A.P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Rowe, H.C., Hansen, B.G., Halkier, B.A. & Kliebenstein, D.J. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20, 1199–1216 (2008).
Joseph, B., Corwin, J.A., Li, B., Atwell, S. & Kliebenstein, D.J. Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2, e00776 (2013).
Fernie, A.R. & Schauer, N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 25, 39–48 (2009).
Traka, M.H. & Mithen, R.F. Plant science and human nutrition: challenges in assessing health-promoting properties of phytochemicals. Plant Cell 23, 2483–2497 (2011).
Kliebenstein, D.J., Gershenzon, J. & Mitchell-Olds, T. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159, 359–370 (2001).
Zhang, H. et al. A core collection and mini core collection of Oryza sativa L. in China. Theor. Appl. Genet. 122, 49–61 (2011).
Yu, S.B. et al. Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theor. Appl. Genet. 108, 131–140 (2003).
Yan, W.G. et al. Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol. Breed. 24, 277–292 (2009).
McNally, K.L. et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. USA 106, 12273–12278 (2009).
Murray, M.G. & Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980).
Wang, J. et al. The diploid genome sequence of an Asian individual. Nature 456, 60–65 (2008).
Xie, W. et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. USA 107, 10578–10583 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Chen, W. et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780 (2013).
Dresen, S., Ferreiros, N., Gnann, H., Zimmermann, R. & Weinmann, W. Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal. Bioanal. Chem. 396, 2425–2434 (2010).
Retief, J.D. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132, 243–258 (2000).
Zhao, K. et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3, e4 (2007).
Li, M.X., Yeung, J.M., Cherny, S.S. & Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131, 747–756 (2012).
Barrett, J.C. Haploview: visualization and analysis of SNP genotype data. Cold Spring Harb. Protoc. 2009, pdb.ip71 (2009).
Zhou, G. et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 109, 15847–15852 (2012).
Yu, H. et al. Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS ONE 6, e17595 (2011).
Zeng, Z.B. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90, 10972–10976 (1993).
Broman, K.W., Wu, H., Sen, S. & Churchill, G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003).
Wang, J. et al. A global analysis of QTLs for expression variations in rice shoots at the early seedling stage. Plant J. 63, 1063–1074 (2010).
Chu, Z. et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255 (2006).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
Livak, K.J. & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
We appreciate the critical reading and helpful comments on the manuscript made by C. Martin from John Innes Centre, UK and Q. Zhang from Huazhong Agricultural University, China. We thank W. Yan for kindly providing 148 varieties from a mini-core subset of the US Department of Agriculture rice gene bank. This work was supported by the Major State Basic Research Development Program of China (973 Program) (number 2013CB127001), the National High Technology R&D Program of China (863 Program) (numbers 2012AA10A303 and 2012AA10A304), the National Natural Science Foundation of China (numbers 31070267 and 31100962) and the Program for New Century Excellent Talents in University of Ministry of Education in China (NCET-09-0401). We are also thankful for support from the Ministry of Science and Technology (numbers 2010CB125901 and 2011CB100304).
Author information
Authors and Affiliations
Contributions
J.L. conceived the project and supervised the study. X. Lian and K.L. prepared the material for genotyping. W.C., Y.G., L.G. and Y.L. performed most of the experiments. W.C., X. Liu, H.Z. and J.L. carried out the metabolite analyses. X. Lian, S.Y., H.D., W.Z., L.Z. and G.W. participated in the material preparation. W.C., W.X., W.W. and J.L. analyzed the data. J.L. wrote the paper. All of the authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1-15 and Supplementary Tables 6, 7 and 23. (PDF 16370 kb)
Supplementary Table 1
Metabolite reporting checklist and recommendations for LC-MS (XLSX 11 kb)
Supplementary Table 2
The (almost) non-redundant MS2T library of rice leaf (XLSX 124 kb)
Supplementary Table 3
Scheduled MRM (multiple reaction monitoring) transitions for widely targeted metabolite analysis in rice leaf (XLSX 147 kb)
Supplementary Table 4
The list of collected 529 rice accessions (XLSX 84 kb)
Supplementary Table 5
Data matrix of 840 metabolites in 524 accessions of rice germplasms (including repeat1 and repeat2) (XLSX 10486 kb)
Supplementary Table 8
The list of total 2,947 significant SNPs detected in at least one of the populations (XLSX 267 kb)
Supplementary Table 9
The list of 634 loci detected in at least one of the populations (XLSX 56 kb)
Supplementary Table 10
The list of 551 lead SNPs that were repeatedly detected (XLSX 110 kb)
Supplementary Table 11
The list of 356 loci that were repeatedly detected (XLSX 77 kb)
Supplementary Table 12
The full lists of significant associations of metabolic GWAS (mGWAS) (XLSX 4949 kb)
Supplementary Table 13
The full lists of significant associations of metabolic GWAS (mGWAS) (XLSX 19291 kb)
Supplementary Table 14
Manhattan plots of 356 loci that were repeatedly detected (XLSX 39 kb)
Supplementary Table 15
Results of analysis of two-locus interactions (XLSX 205 kb)
Supplementary Table 16
Statistics of significant loci on the chromosomes (XLSX 16 kb)
Supplementary Table 17
The list of significant loci detected in indica and japonica subspecies (XLSX 58 kb)
Supplementary Table 18
The list of significant loci detected for subspecies differentiation metabolites in indica and japonica subspecies (XLSX 21 kb)
Supplementary Table 19
The full list of identified or annotated metabolites that were supported by GWAS (XLSX 40 kb)
Supplementary Table 20
The full list of candidate genes (XLSX 20 kb)
Supplementary Table 21
Metabolic profiling of transgenic lines overexpressing the candidate genes (XLSX 47 kb)
Supplementary Table 22
The results of overlap between GWAS loci and mQTL (XLSX 24 kb)
Rights and permissions
About this article
Cite this article
Chen, W., Gao, Y., Xie, W. et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46, 714–721 (2014). https://doi.org/10.1038/ng.3007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3007
This article is cited by
-
Utilizing transcriptomics and metabolomics to unravel key genes and metabolites of maize seedlings in response to drought stress
BMC Plant Biology (2024)
-
Integrated transcriptome and metabolome analysis revealed that HaMYB1 modulates anthocyanin accumulation to deepen sunflower flower color
Plant Cell Reports (2024)
-
Common and specific genetic basis of metabolite-mediated drought responses in rice
Stress Biology (2024)
-
Metabolic GWAS-based dissection of genetic basis underlying nutrient quality variation and domestication of cassava storage root
Genome Biology (2023)
-
Characterization of novel loci controlling seed oil content in Brassica napus by marker metabolite-based multi-omics analysis
Genome Biology (2023)