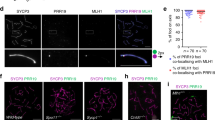
Abstract
Crossover recombination facilitates the accurate segregation of homologous chromosomes during meiosis1,2. In mammals, poorly characterized regulatory processes ensure that every pair of chromosomes obtains at least one crossover, even though most recombination sites yield non-crossovers3. Designation of crossovers involves selective localization of the SUMO ligase RNF212 to a minority of recombination sites, where it stabilizes pertinent factors such as MutSγ (ref. 4). Here we show that the ubiquitin ligase HEI10 (also called CCNB1IP1)5,6 is essential for this crossover/non-crossover differentiation process. In HEI10-deficient mice, RNF212 localizes to most recombination sites, and dissociation of both RNF212 and MutSγ from chromosomes is blocked. Consequently, recombination is impeded, and crossing over fails. In wild-type mice, HEI10 accumulates at designated crossover sites, suggesting that it also has a late role in implementing crossing over. As with RNF212, dosage sensitivity for HEI10 indicates that it is a limiting factor for crossing over. We suggest that SUMO and ubiquitin have antagonistic roles during meiotic recombination that are balanced to effect differential stabilization of recombination factors at crossover and non-crossover sites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sakuno, T., Tanaka, K., Hauf, S. & Watanabe, Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev. Cell 21, 534–545 (2011).
Hirose, Y. et al. Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet. 7, e1001329 (2011).
Jones, G.H. The control of chiasma distribution. Symp. Soc. Exp. Biol. 38, 293–320 (1984).
Reynolds, A. et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013).
Toby, G.G., Gherraby, W., Coleman, T.R. & Golemis, E.A. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol. Cell Biol. 23, 2109–2122 (2003).
Ward, J.O. et al. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, e139 (2007).
Kong, A. et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319, 1398–1401 (2008).
Fledel-Alon, A. et al. Variation in human recombination rates and its genetic determinants. PLoS ONE 6, e20321 (2011).
Chowdhury, R., Bois, P.R., Feingold, E., Sherman, S.L. & Cheung, V.G. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5, e1000648 (2009).
Kong, A. et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 46, 11–16 (2014).
Cheng, C.H. et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20, 2067–2081 (2006).
Strong, E.R. & Schimenti, J.C. Evidence implicating CCNB1IP1, a RING domain–containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes (Basel) 1, 440–451 (2010).
Kolas, N.K. & Cohen, P.E. Novel and diverse functions of the DNA mismatch repair family in mammalian meiosis and recombination. Cytogenet. Genome Res. 107, 216–231 (2004).
Snowden, T., Acharya, S., Butz, C., Berardini, M. & Fishel, R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451 (2004).
Hunter, N. in Molecular Genetics of Recombination (eds. Aguilera, A. & Rothstein, R.) 381–442 (Springer-Verlag, Heidelberg, 2006).
Carlton, P.M. Three-dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res. 16, 351–365 (2008).
Chicheportiche, A., Bernardino-Sgherri, J., de Massy, B. & Dutrillaux, B. Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J. Cell Sci. 120, 1733–1742 (2007).
Fernandez-Capetillo, O. et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4, 497–508 (2003).
Baudat, F., Manova, K., Yuen, J.P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Romanienko, P.J. & Camerini-Otero, R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000).
de Vries, F.A. et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 (2005).
Lipkin, S.M. et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31, 385–390 (2002).
Eaker, S., Cobb, J., Pyle, A. & Handel, M.A. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev. Biol. 249, 85–95 (2002).
Holloway, J.K., Booth, J., Edelmann, W., McGowan, C.H. & Cohen, P.E. MUS81 generates a subset of MLH1-MLH3–independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (2008).
Qiao, H., Lohmiller, L. & Anderson, L. Cohesin proteins load sequentially during prophase I in tomato primary microsporocytes. Chromosome Res. 19, 193–207 (2011).
Costa, Y. et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118, 2755–2762 (2005).
Cobb, J., Cargile, B. & Handel, M.A. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev. Biol. 205, 49–64 (1999).
Holloway, J.K., Morelli, M.A., Borst, P.L. & Cohen, P.E. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188, 779–789 (2010).
Acknowledgements
We thank A. Kong for communicating unpublished results. This work was supported by US National Institutes of Health (NIH) grants R01GM084955 to N.H., R01GM45415 to J.S. and HD041012 to P.E.C. and by National Science Foundation grant CAREER 0844941 to J.W. N.H. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
H.Q., H.B.D.P.R., Y.Y., J.W. and N.H. conceived and designed the experiments. H.Q., H.B.D.P.R., Y.Y., J.H.F., J.M.C., D.C.D., K.E.N., R.K.S., E.S., J.K.H., J.W. and N.H. performed the experiments. H.Q., H.B.D.P.R., Y.Y., J.H.F., J.W. and N.H. analyzed the data. J.S., E.S. and P.E.C. contributed reagents, materials and/or analysis tools. H.Q., H.B.D.P.R., Y.Y., J.W. and N.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 8260 kb)
Rights and permissions
About this article
Cite this article
Qiao, H., Prasada Rao, H., Yang, Y. et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet 46, 194–199 (2014). https://doi.org/10.1038/ng.2858
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2858
This article is cited by
-
The RING Domain of Rice HEI10 is Essential for Male, But Not Female Fertility
Rice (2024)
-
RNA polymerase II pausing is essential during spermatogenesis for appropriate gene expression and completion of meiosis
Nature Communications (2024)
-
A novel recombination protein C12ORF40/REDIC1 is required for meiotic crossover formation
Cell Discovery (2023)
-
Signalling mechanisms and cellular functions of SUMO
Nature Reviews Molecular Cell Biology (2022)
-
Joint control of meiotic crossover patterning by the synaptonemal complex and HEI10 dosage
Nature Communications (2022)