Abstract
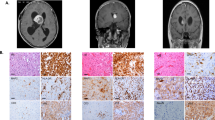
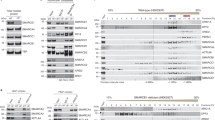
One-third of all primary central nervous system tumors in adults are meningiomas1. Rarely, meningiomas occur at multiple sites, usually occurring in individuals with type 2 neurofibromatosis (NF2). We sequenced the exomes of three unrelated individuals with familial multiple spinal meningiomas without NF2 mutations. We identified two individuals with heterozygous loss-of-function mutations in the SWI/SNF chromatin-remodeling complex subunit gene SMARCE1. Sequencing of SMARCE1 in six further individuals with spinal meningiomas identified two additional heterozygous loss-of-function mutations. Tumors from individuals with SMARCE1 mutations were of clear-cell histological subtype, and all had loss of SMARCE1 protein, consistent with a tumor suppressor mechanism. Our findings identify multiple-spinal-meningioma disease as a new discrete entity and establish a key role for the SWI/SNF complex in the pathogenesis of both meningiomas and tumors with clear-cell histology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
CBTRUS. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004–2008 (Central Brain Tumor Registry of the United States, Hinsdale, Illinois, 2012).
Bondy, M. & Ligon, B.L. Epidemiology and etiology of intracranial meningiomas: a review. J. Neurooncol. 29, 197–205 (1996).
Smith, M.J. et al. Cranial meningiomas in 411 neurofibromatosis type 2 (NF2) patients with proven gene mutations: clear positional effect of mutations, but absence of female severity effect on age at onset. J. Med. Genet. 48, 261–265 (2011).
Ruttledge, M.H. et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat. Genet. 6, 180–184 (1994).
Antinheimo, J. et al. Population-based analysis of sporadic and type 2 neurofibromatosis–associated meningiomas and schwannomas. Neurology 54, 71–76 (2000).
Evans, D.G., Watson, C., King, A., Wallace, A.J. & Baser, M.E. Multiple meningiomas: differential involvement of the NF2 gene in children and adults. J. Med. Genet. 42, 45–48 (2005).
Bacci, C. et al. Schwannomatosis associated with multiple meningiomas due to a familial SMARCB1 mutation. Neurogenetics 11, 73–80 (2010).
Christiaans, I. et al. Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J. Med. Genet. 48, 93–97 (2011).
van den Munckhof, P., Christiaans, I., Kenter, S.B., Baas, F. & Hulsebos, T.J. Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the falx cerebri. Neurogenetics 13, 1–7 (2012).
Rieske, P. et al. Molecular heterogeneity of meningioma with INI1 mutation. Mol. Pathol. 56, 299–301 (2003).
Schmitz, U. et al. INI1 mutations in meningiomas at a potential hotspot in exon 9. Br. J. Cancer 84, 199–201 (2001).
Hadfield, K.D., Smith, M.J., Trump, D., Newman, W.G. & Evans, D.G. SMARCB1 mutations are not a common cause of multiple meningiomas. J. Med. Genet. 47, 567–568 (2010).
Shen, Y. et al. Genomic profiling distinguishes familial multiple and sporadic multiple meningiomas. BMC Med. Genomics 2, 42 (2009).
Hemminki, K. & Li, X. Familial risks in nervous system tumors. Cancer Epidemiol. Biomarkers Prev. 12, 1137–1142 (2003).
Dobbins, S.E. et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat. Genet. 43, 825–827 (2011).
Chi, T.H. et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195–199 (2002).
Mitchell, R.J., Farrington, S.M., Dunlop, M.G. & Campbell, H. Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am. J. Epidemiol. 156, 885–902 (2002).
Wang, W. et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA 95, 492–498 (1998).
Kazantseva, A. et al. N-terminally truncated BAF57 isoforms contribute to the diversity of SWI/SNF complexes in neurons. J. Neurochem. 109, 807–818 (2009).
Wang, L. et al. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol. Cell Biol. 25, 7953–7965 (2005).
García-Pedrero, J.M., Kiskinis, E., Parker, M.G. & Belandia, B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J. Biol. Chem. 281, 22656–22664 (2006).
Belandia, B., Orford, R.L., Hurst, H.C. & Parker, M.G. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 21, 4094–4103 (2002).
Kiskinis, E., Garcia-Pedrero, J.M., Villaronga, M.A., Parker, M.G. & Belandia, B. Identification of BAF57 mutations in human breast cancer cell lines. Breast Cancer Res. Treat. 98, 191–198 (2006).
Luo, B. et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. USA 105, 20380–20385 (2008).
Villaronga, M.A. et al. Identification and characterization of novel potentially oncogenic mutations in the human BAF57 gene in a breast cancer patient. Breast Cancer Res. Treat. 128, 891–898 (2011).
Jagani, Z. et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat. Med. 16, 1429–1433 (2010).
Aavikko, M. et al. Loss of SUFU function in familial multiple meningioma. Am. J. Hum. Genet. 91, 520–526 (2012).
Kuratsu, J., Kochi, M. & Ushio, Y. Incidence and clinical features of asymptomatic meningiomas. J. Neurosurg. 92, 766–770 (2000).
Wiegand, K.C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Tsurusaki, Y. et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat. Genet. 44, 376–378 (2012).
Santen, G.W. et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat. Genet. 44, 379–380 (2012).
Wong, D.R., Beneke, J.S., Janus, S.C. & Levine, S.C. Coffin-Siris syndrome and neurofibromatosis type 2: a clinicopathologic enigma. Laryngoscope 119, S129 (2009).
Acknowledgements
We thank the families for their contributions to this study. We also thank N. Bowers and A. Wallace for NF2 mutation screening and T. Gledhill for immunohistochemistry images. M.J.S. is supported by a Young Investigator Award (2011-01-006) from the Children's Tumor Foundation. We also acknowledge support from the Association for International Cancer Research (12-0275) and the Manchester Biomedical Research Centre. D.G.E. is a National Institute for Health Research (NIHR) Senior Clinical Investigator. Finally, we thank the NHLBI GO Exome Sequencing Project and its ongoing studies, which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the Women's Health Initiative (WHI) Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010).
Author information
Authors and Affiliations
Contributions
M.J.S., W.G.N. and D.G.E. designed the study and wrote the manuscript. J.O. and S.S.B. performed exome sequencing and bioinformatics analysis. M.J.S. and K.D.H. performed Sanger sequencing. D.G.E., G.P., D.F., S.S., D.R., D.E. and J.C. provided detailed clinical information and samples for analysis. D.d.P. performed histopathological review and immunohistochemistry studies. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 (PDF 762 kb)
Rights and permissions
About this article
Cite this article
Smith, M., O'Sullivan, J., Bhaskar, S. et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet 45, 295–298 (2013). https://doi.org/10.1038/ng.2552
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2552
This article is cited by
-
Lumbar clear cell meningioma mimicking schwannoma 7 years after resection of the same type of intracranial tumor: a case report
Journal of Medical Case Reports (2024)
-
A novel BRAF::PTPRN2 fusion in meningioma: a case report
Acta Neuropathologica Communications (2023)
-
A novel patient-derived meningioma spheroid model as a tool to study and treat epithelial-to-mesenchymal transition (EMT) in meningiomas
Acta Neuropathologica Communications (2023)
-
Review of meningioma diagnosis and management
Egyptian Journal of Neurosurgery (2023)
-
A current review of spinal meningiomas: epidemiology, clinical presentation and management
Journal of Neuro-Oncology (2023)