Abstract
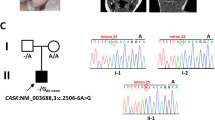
De novo somatic mutations in focal areas are well documented in diseases such as neoplasia but are rarely reported in malformation of the developing brain. Hemimegalencephaly (HME) is characterized by overgrowth of either one of the two cerebral hemispheres. The molecular etiology of HME remains a mystery. The intractable epilepsy that is associated with HME can be relieved by the surgical treatment hemispherectomy, allowing sampling of diseased tissue. Exome sequencing and mass spectrometry analysis in paired brain-blood samples from individuals with HME (n = 20 cases) identified de novo somatic mutations in 30% of affected individuals in the PIK3CA, AKT3 and MTOR genes. A recurrent PIK3CA c.1633G>A mutation was found in four separate cases. Identified mutations were present in 8–40% of sequenced alleles in various brain regions and were associated with increased neuronal S6 protein phosphorylation in the brains of affected individuals, indicating aberrant activation of mammalian target of rapamycin (mTOR) signaling. Thus HME is probably a genetically mosaic disease caused by gain of function in phosphatidylinositol 3-kinase (PI3K)-AKT3-mTOR signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Blümcke, I. et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia 52, 158–174 (2011).
Barkovich, A.J., Guerrini, R., Kuzniecky, R.I., Jackson, G.D. & Dobyns, W.B. A developmental and genetic classification for malformations of cortical development: update 2012. Brain 135, 1348–1369 (2012).
Salamon, N. et al. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain 129, 352–365 (2006).
Di Rocco, C., Battaglia, D., Pietrini, D., Piastra, M. & Massimi, L. Hemimegalencephaly: clinical implications and surgical treatment. Childs Nerv. Syst. 22, 852–866 (2006).
Crino, P.B. mTOR: a pathogenic signaling pathway in developmental brain malformations. Trends Mol. Med. 17, 734–742 (2011).
Palmini, A. et al. Terminology and classification of the cortical dysplasias. Neurology 62, S2–S8 (2004).
Jozwiak, J., Jozwiak, S. & Skopinski, P. Immunohistochemical and microscopic studies on giant cells in tuberous sclerosis. Histol. Histopathol. 20, 1321–1326 (2005).
Boer, K. et al. A neuropathological study of two autopsy cases of syndromic hemimegalencephaly. Neuropathol. Appl. Neurobiol. 33, 455–470 (2007).
Yu, J. et al. Targeted gene expression analysis in hemimegalencephaly: activation of beta-catenin signaling. Brain Pathol. 15, 179–186 (2005).
Griffiths, P.D., Welch, R.J., Gardner-Medwin, D., Gholkar, A. & McAllister, V. The radiological features of hemimegalencephaly including three cases associated with proteus syndrome. Neuropediatrics 25, 140–144 (1994).
Lindhurst, M.J. et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 365, 611–619 (2011).
Mefford, H.C. et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann. Neurol. 70, 974–985 (2011).
Alkan, C., Coe, B.P. & Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 (2011).
Iafrate, A.J. et al. Detection of large-scale variation in the human genome. Nat. Genet. 36, 949–951 (2004).
Jonkman, M.F. & Pasmooij, A.M. Revertant mosaicism—patchwork in the skin. N. Engl. J. Med. 360, 1680–1682 (2009).
Choate, K.A. et al. Mitotic recombination in patients with ichthyosis causes reversion of dominant mutations in KRT10. Science 330, 94–97 (2010).
Jongmans, M.C. et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am. J. Hum. Genet. 90, 426–433 (2012).
Merks, J.H. et al. PTEN hamartoma tumour syndrome: variability of an entity. J. Med. Genet. 40, e111 (2003).
Roth, A. et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next generation sequencing data. Bioinformatics 28, 907–913 (2012).
Thomas, R.K. et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 39, 347–351 (2007).
Oeth, P., del Mistro, G., Marnellos, G., Shi, T. & van den Boom, D. Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY). Methods Mol. Biol. 578, 307–343 (2009).
Tschopp, O. et al. Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132, 2943–2954 (2005).
Easton, R.M. et al. Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol. Cell. Biol. 25, 1869–1878 (2005).
Tokuda, S. et al. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Hum. Mol. Genet. 20, 988–999 (2011).
Broderick, D.K. et al. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 64, 5048–5050 (2004).
Samuels, Y. et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 304, 554 (2004).
Kang, S., Bader, A.G. & Vogt, P.K. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc. Natl. Acad. Sci. USA 102, 802–807 (2005).
Davies, M.A. et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer 99, 1265–1268 (2008).
Sato, T., Nakashima, A., Guo, L., Coffman, K. & Tamanoi, F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene 29, 2746–2752 (2010).
Hardt, M., Chantaravisoot, N. & Tamanoi, F. Activating mutations of TOR (target of rapamycin). Genes Cells 16, 141–151 (2011).
Gao, X. & Pan, D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15, 1383–1392 (2001).
Potter, C.J., Huang, H. & Xu, T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105, 357–368 (2001).
Tapon, N., Ito, N., Dickson, B.J., Treisman, J.E. & Hariharan, I.K. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105, 345–355 (2001).
Ito, N. & Rubin, G.M. gigas, a Drosophila homolog of tuberous sclerosis gene product–2, regulates the cell cycle. Cell 96, 529–539 (1999).
Bissler, J.J. et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N. Engl. J. Med. 358, 140–151 (2008).
Sharma, S., Sankhyan, N., Kabra, M. & Kumar, A. Hypomelanosis of Ito with hemimegalencephaly. Dermatol. Online J. 15, 12 (2009).
Chapman, K. & Cardenas, J.F. Hemimegalencephaly in a patient with a neurocutaneous syndrome. Semin. Pediatr. Neurol. 15, 190–193 (2008).
Auriemma, A. et al. Hemimegalencephaly in hypomelanosis of Ito: early sonographic pattern and peculiar MR findings in a newborn. Eur. J. Ultrasound 12, 61–67 (2000).
Hemb, M. et al. Improved outcomes in pediatric epilepsy surgery: the UCLA experience, 1986–2008. Neurology 74, 1768–1775 (2010).
Jonas, R. et al. Cerebral hemispherectomy: hospital course, seizure, developmental, language, and motor outcomes. Neurology 62, 1712–1721 (2004).
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011).
Acknowledgements
We thank J. Meerloo at the UCSD Neurosciences Core for microscopy services (supported by the US National Institute of Neurological Disorders and Stroke; P30NS047101), the Genomics Core at Cedars-Sinai Medical Center, the Broad Institute (supported by the US National Human Genome Research Institute; U54HG003067 to E. Lander) for sequencing support and analysis, D. Neelam (Sequenom) for technical support and W.B. Dobyns and M.L. Warman for sharing unpublished results. This work was supported by grants from the Daland Fellowship from the American Philosophical Society (to J.H.L.), the US National Institutes of Health (R01 NS038992 to G.W.M. and R01 NS048453, R01 NS052455, R01 NS041537 and P01 HD070494), the Simons Foundation Autism Research Initiative and the Howard Hughes Medical Institute (to J.G.G.).
Author information
Authors and Affiliations
Contributions
J.H.L. and J.L.S. organized the project and performed genetic studies. K.J.H., A.C. and J.H.L. recruited subjects. J.H.L., M.H. and T.D.-S. performed immunostaining. C.R. and S.B.G. generated and interpreted exome results. S.K., A.H., E.S., V.B. and J.H.L. performed JointSNVMix analysis. V.F. and J.H.L. oversaw SNP genotyping and analyzed CNVs. G.W.M. performed surgeries and managed samples along with M.H. G.W.M. and J.G.G. conceived of the project and oversaw data collection and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1 and Supplementary Figure 1–3 (PDF 1515 kb)
Rights and permissions
About this article
Cite this article
Lee, J., Huynh, M., Silhavy, J. et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 44, 941–945 (2012). https://doi.org/10.1038/ng.2329
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2329
This article is cited by
-
Clinical characteristics and surgical management of facial infiltrating lipomatosis: a single center experience
Head & Face Medicine (2024)
-
Delineation of the phenotypes and genotypes of facial infiltrating lipomatosis associated with PIK3CA mutations
Orphanet Journal of Rare Diseases (2023)
-
The specific DNA methylation landscape in focal cortical dysplasia ILAE type 3D
Acta Neuropathologica Communications (2023)
-
Smith-Kingsmore syndrome with nystagmus as the initial symptom
Acta Epileptologica (2023)
-
Cytogenomic epileptology
Molecular Cytogenetics (2023)