Abstract
Adjuvant chemotherapy improves survival of patients with stage I–III breast cancer but it is being increasingly recognized that the benefit is not equal for all patients. Molecular characteristics of the cancer affect sensitivity to chemotherapy. In general, estrogen-receptor-negative disease is more sensitive to chemotherapy than estrogren-receptor-positive disease. Large-scale genomic analyses of breast cancer suggest that further molecular subsets may exist within the categories defined by hormone receptor status. It is hoped that the new molecular classification schemes might improve patient selection for therapy. Before any new molecular classification (or predictive test) is adopted for routine clinical use, however, several criteria need to be met. There must be an agreed and reproducible method by which to assign molecular class to a new case. Cancers that belong to different molecular classes must show differences in disease outcome and treatment efficacy that affect management and treatment selection. Also desirable are results from prospective clinical trials that demonstrate improved patient outcome when the new test is used in decision-making, compared with the current standard of care. This Review describes the current limitations and future promises of gene-expression-based molecular classification of breast cancer and how it might impact on selection of adjuvant therapy for individual patients.
Key Points
-
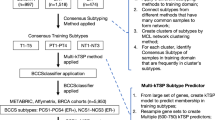
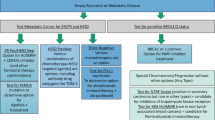
Gene expression profiling of breast cancer has revealed large-scale molecular differences between ER-positive, ER-negative and HER2-amplified cancers
-
It is more appropriate to think of breast cancer as at least two to three distinct diseases than as a single disease with heterogeneous ER and HER expression
-
Molecular classification of breast cancer provides a new framework for the study of breast cancer, but how many robust molecular subtypes exist and how best to assign a molecular class to new cases is currently unknown; standard methods for molecular class determination are needed
-
Multigene signatures can be used to help guide therapy and predict prognosis and response to preoperative chemotherapy
-
The extent to which multigene signatures improve patient outcome compared with current clinicopathologic variable-based predictions is yet to be determined in prospective clinical trials
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Early Breast Cancer Trialists' Collaborative Group (EBCTCG) (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365: 1687–1717
Campone M et al. (2005) Secondary leukemia after epirubicin-based adjuvant chemotherapy in operable breast cancer patients: 16 years experience of the French Adjuvant Study Group. Ann Oncol 16: 1343–1351
Doyle JJ et al. (2005) Chemotherapy and cardiotoxicity in older breast cancer patients: a population-based study. J Clin Oncol 23: 8597–8605
Trudeau M et al. (2005) Selection of adjuvant chemotherapy for treatment of node-positive breast cancer. Lancet Oncol 6: 886–898
Nowak AK et al. (2004) Systematic review of taxane-containing versus non-taxane-containing regimens for adjuvant and neoadjuvant treatment of early breast cancer. Lancet Oncol 5: 372–380
Bast RC Jr et al. (2001) 2000 Update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol 19: 1865–1878
D'Eredita G et al. (2001) Prognostic factors in breast cancer: the predictive value of the Nottingham Prognostic Index in patients with a long-term follow-up that were treated in a single institution. Eur J Cancer 37: 591–596
Olivotto IA et al. (2005) Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol 23: 2716–2725
Rouzier R et al. (2005) Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol 23: 8331–8339
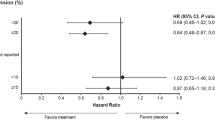
Berry DA et al. (2006) Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 295: 1658–1667
Guarneri V et al. (2006) Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol 24: 1037–1044
Bear HD et al. (2003) The effect on tumor response of adding sequential preoperative docetaxel to preoperative doxorubicin and cyclophosphamide: preliminary results from National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol 21: 4165–4174
Pusztai L et al. (2003) Gene expression profiles obtained from single passage fine needle aspirations (FNA) of breast cancer reliably identify prognostic/predictive markers such as estrogen (ER) and HER-2 receptor status and reveal large scale molecular differences between ER-negative and ER-positive tumors. Clin Cancer Res 9: 2406–2415
Gruvberger S et al. (2001) Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res 61: 5979–5984
Rhodes A et al. (2000) Reliability of immunohistochemical demonstration of oestrogen receptors in routine practice: interlaboratory variance in the sensitivity of detection and evaluation of scoring systems. J Clin Pathol 53: 125–130
Bonneterre J et al. (2000) Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: results of the Tamoxifen or Arimidex Randomized Group Efficacy and Tolerability Study. J Clin Oncol 18: 3748–3757
Mouridsen H et al. (2001) Superior efficacy of letrozole versus tamoxifen as first-line therapy for postmenopausal women with advanced breast cancer: results of a phase III study of the International Letrozole Breast Cancer Group. J Clin Oncol 19: 2596–2606
Lacroix M et al. (2001) Estrogen receptor analysis in primary breast tumors by ligand-binding assay, immunocytochemical assay, and northern blot: a comparison. Breast Cancer Res Treat 67: 263–271
Paik S et al. (2004) A multi gene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826
Habel LA et al. (2005) Gene expression and breast cancer mortality in Northern California Kaiser Permanente patients: a large population-based case control study [abstract]. Proc Am Soc Clin Oncol 24: 603a
Paik S et al. (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24: 3726–3734
Ma XJ et al. (2004) A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 5: 607–616
Jansen MP et al. (2005) Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J Clin Oncol 23: 732–740
Symmans WF et al. (2005) Measurements of estrogen receptor and reporter genes from micro arrays determine receptor status and time to recurrence following adjuvant tamoxifen therapy [abstract]. Breast Cancer Res Treat 94 (Suppl 1): S308a
Loi S et al. (2005) Prediction of early relapses on tamoxifen in early-stage breast cancer (BC): a potential tool for adjuvant aromatase inhibitor (AI) tailoring [abstract #509]. Proc Am Soc Clin Oncol
Radmacher MD et al. (2002) A paradigm for class prediction using gene expression profiles. J Comput Biol 9: 505–511
Perou CM et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752
Sorlie T et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874
Sorlie T et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423
Hu Z et al. (2006) The molecular portraits of breast tumors are conserved across microarray platfroms. BMC Genomics 7: 96
Sotiriou C et al. (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA 100: 10393–10398
Pusztai L et al. (2003) Gene expression profiles obtained from single passage fine needle aspirations (FNA) of breast cancer reliably identify prognostic/predictive markers such as estrogen (ER) and HER-2 receptor status and reveal large scale molecular differences between ER-negative and ER-positive tumors. Clin Cancer Res 9: 2406–2415
Rouzier R et al. (2005) Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res 11: 5678–5685
McShane LM et al. (2002) Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics 18: 1462–1469
Milligan GW and Cooper MC (1985) An examination of procedures for determining the number of clusters in a data set. Psychometrika 50: 159–179
Tibshirani R et al. (2001) Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc B 63: 411–423
Nielsen TO et al. (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374
Abd El-Rehim DM et al. (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116: 340–350
Van de Rijn M et al. (2002) Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol 161: 1991–1996
Radmacher MD et al. (2002) A paradigm for class prediction using gene expression profiles. J Comput Biol 9: 505–511
van't Veer LJ et al. (2001) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536
van de Vijver MJ et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009
Buyse M et al. (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98: 1183–1192
Wang Y et al. (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365: 671–679
Foekens JA et al. (2006) Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol 24: 1665–1671
Chang JC et al. (2003) Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 362: 362–369
Iwao-Koizumi K et al. (2005) Prediction of docetaxel response in human breast cancer by gene expression profiling. J Clin Oncol 23: 422–431
Hess KR et al. (2006) Pharmacogenomic predictor of sensitivity to preoperative paclitaxel and 5-fluorouracil, doxorubicin, cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 24: 4236–4244
Ayers M et al. (2004) Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 22: 2284–2293
Gianni L et al. (2005) Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol 23: 7265–7277
Yoshimoto M et al. (2004) Prediction of the therapeutic response to paclitaxel by gene expression profiling in neoadjuvant chemotherapy for breast cancer. 40th Annual ASCO Meeting Proceeding [abstract #500]. J Clin Oncol 22 (Suppl): a14S
Folgueira MA et al. (2005) Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin Cancer Res 11: 7434–7443
Modlich O et al. (2005) Predictors of primary breast cancers responsiveness to preoperative epirubicin/cyclophosphamide-based chemotherapy: translation of microarray data into clinically useful predictive signatures. J Translational Med 3: 32
Petit T et al. (2004) Comparative value of tumour grade, hormonal receptors, Ki-67, HER-2 and topoisomerase II alpha status as predictive markers in breast cancer patients treated with neoadjuvant anthracycline-based chemotherapy. Eur J Cancer 40: 205–211
Di Leo A et al. (2002) HER-2 amplification and topoisomerase IIalpha gene aberrations as predictive markers in node-positive breast cancer patients randomly treated either with an anthracycline-based therapy or with cyclophosphamide, methotrexate, and 5-fluorouracil. Clin Cancer Res 8: 1107–1116
Press MF et al. (2005) Topoisomerase II-alpha gene amplification as a predictor of responsiveness to anthracycline-containing chemotherapy in the Breast Cancer International Research Group 006 clinical trial of trastuzumab (Herceptin) in the adjuvant setting [abstract #1045]. Breast Cancer Res Treat 94 (Suppl 1): S32
Fedier A et al. (2003) p53-deficient cells display increased sensitivity to anthracyclines after loss of the catalytic subunit of the DNA-dependent protein kinase. Int J Oncol 23: 1431–1437
Geisler S et al. (2001) Influence of TP53 gene alterations and c-erbB-2 expression on the response to treatment with doxorubicin in locally advanced breast cancer. Cancer Res 61: 2505–2512
Bertheau P et al. (2002) Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet 360: 852–854
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
Related links in Nature Research
Recurrence score can predict chemotherapy benefit for breast cancer patients
Gene expression profiling for individualized breast cancer chemotherapy: success or not?
Rights and permissions
About this article
Cite this article
Andre, F., Pusztai, L. Molecular classification of breast cancer: implications for selection of adjuvant chemotherapy. Nat Rev Clin Oncol 3, 621–632 (2006). https://doi.org/10.1038/ncponc0636
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/ncponc0636
This article is cited by
-
Molecular Biology in the Breast Clinics—Current status and future perspectives
Indian Journal of Surgical Oncology (2021)
-
Recent advances in nanomaterials-based electrochemical immunosensors and aptasensors for HER2 assessment in breast cancer
Microchimica Acta (2021)
-
Breast cancer risk in premalignant lesions: osteopontin splice variants indicate prognosis
British Journal of Cancer (2018)
-
Etiology, pathophysiology and biomarkers of interstitial cystitis/painful bladder syndrome
Archives of Gynecology and Obstetrics (2017)
-
Serum sonic hedgehog (SHH) and interleukin-(IL-6) as dual prognostic biomarkers in progressive metastatic breast cancer
Scientific Reports (2017)