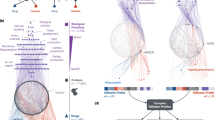
Abstract
Although it is increasingly being recognized that drug-target interaction networks can be powerful tools for the interrogation of systems biology and the rational design of multitargeted drugs, there is no generalized, statistically validated approach to harmonizing sequence-dependent and pharmacology-dependent networks. Here we demonstrate the creation of a comprehensive kinome interaction network based not only on sequence comparisons but also on multiple pharmacology parameters derived from activity profiling data. The framework described for statistical interpretation of these network connections also enables rigorous investigation of chemotype-specific interaction networks, which is critical for multitargeted drug design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Roth, B.L., Sheffler, D.J. & Kroeze, W.K. Nat. Rev. Drug Discov. 3, 353–359 (2004).
Frantz, S. Nature 437, 942–943 (2005).
Morphy, R. J. Med. Chem. 53, 1413–1437 (2010).
Mestres, J., Gregori-Puigjane, E., Valverde, S. & Sole, R.V. Mol. Biosyst. 5, 1051–1057 (2009).
Paolini, G.V., Shapland, R.H., van Hoorn, W.P., Mason, J.S. & Hopkins, A.L. Nat. Biotechnol. 24, 805–815 (2006).
Keiser, M.J. et al. Nat. Biotechnol. 25, 197–206 (2007).
Keiser, M.J. et al. Nature 462, 175–181 (2009).
Oprea, T.I., Tropsha, A., Faulon, J.L. & Rintoul, M.D. Nat. Chem. Biol. 3, 447–450 (2007).
Vogt, I. & Mestres, J. Mol. Inf. 29, 10–14 (2010).
Metz, J.T. & Hajduk, P.J. Curr. Opin. Chem. Biol. 14, 498–504 (2010).
Vieth, M. et al. Biochim. Biophys. Acta 1697, 243–257 (2004).
Vieth, M., Sutherland, J.J., Robertson, D.H. & Campbell, R.M. Drug Discov. Today 10, 839–846 (2005).
Bamborough, P., Drewry, D., Harper, G., Smith, G.K. & Schneider, K. J. Med. Chem. 51, 7898–7914 (2008).
Mestres, J., Gregori-Puigjane, E., Valverde, S. & Sole, R.V. Nat. Biotechnol. 26, 983–984 (2008).
Godden, J.W. & Bajorath, J. J. Mol. Graph. Model. 18, 73–76 (2000).
Godden, J.W. & Bajorath, J. QSAR Comb. Sci. 22, 487–497 (2003).
Author information
Authors and Affiliations
Contributions
J.T.M. performed all of the statistical analyses and created the network visualizations; E.F.J. designed the initial kinome screening panel and supervised the enzymology; N.B.S., P.J.M. and L.K. performed all kinase assays; P.J.H. supervised the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are currently employed by and obtain their salary from Abbott Laboratories.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–6 and Supplementary Tables 1 & 2 descriptions (PDF 1153 kb)
Supplementary Table 1
Kinome pKi data with clusters (xls) (XLS 3912 kb)
Supplementary Table 2
Kinome pharmacology interactions (csv) (CSV 3218 kb)
Supplementary Dataset
Kinome pharmacology analysis, .txt file for Pipeline Pilot (XML 557 kb)
Rights and permissions
About this article
Cite this article
Metz, J., Johnson, E., Soni, N. et al. Navigating the kinome. Nat Chem Biol 7, 200–202 (2011). https://doi.org/10.1038/nchembio.530
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.530
This article is cited by
-
Drug–target affinity prediction with extended graph learning-convolutional networks
BMC Bioinformatics (2024)
-
Integration of fingerprint-based similarity searching and kernel-based partial least squares analysis to predict inhibitory activity against CSK, HER2, JAK1, JAK2, and JAK3
Molecular Diversity (2023)
-
Deep learning in drug discovery: an integrative review and future challenges
Artificial Intelligence Review (2023)
-
Identification of pan-kinase-family inhibitors using graph convolutional networks to reveal family-sensitive pre-moieties
BMC Bioinformatics (2022)
-
Exploring kinase family inhibitors and their moiety preferences using deep SHapley additive exPlanations
BMC Bioinformatics (2022)