Abstract
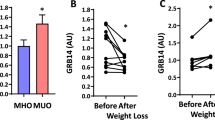
Obesity-associated insulin resistance plays a central role in type 2 diabetes. As such, tyrosine phosphatases that dephosphorylate the insulin receptor (IR) are potential therapeutic targets. The low-molecular-weight protein tyrosine phosphatase (LMPTP) is a proposed IR phosphatase, yet its role in insulin signaling in vivo has not been defined. Here we show that global and liver-specific LMPTP deletion protects mice from high-fat diet-induced diabetes without affecting body weight. To examine the role of the catalytic activity of LMPTP, we developed a small-molecule inhibitor with a novel uncompetitive mechanism, a unique binding site at the opening of the catalytic pocket, and an exquisite selectivity over other phosphatases. This inhibitor is orally bioavailable, and it increases liver IR phosphorylation in vivo and reverses high-fat diet-induced diabetes. Our findings suggest that LMPTP is a key promoter of insulin resistance and that LMPTP inhibitors would be beneficial for treating type 2 diabetes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Referenced accessions
GenBank/EMBL/DDBJ
Protein Data Bank
References
Biddinger, S.B. & Kahn, C.R. From mice to men: insights into the insulin resistance syndromes. Annu. Rev. Physiol. 68, 123–158 (2006).
Kahn, S.E., Hull, R.L. & Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444, 840–846 (2006).
Saltiel, A.R. & Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 (2001).
White, M.F., Shoelson, S.E., Keutmann, H. & Kahn, C.R. A cascade of tyrosine autophosphorylation in the beta-subunit activates the phosphotransferase of the insulin receptor. J. Biol. Chem. 263, 2969–2980 (1988).
Musi, N. & Goodyear, L.J. Insulin resistance and improvements in signal transduction. Endocrine 29, 73–80 (2006).
Alonso, A. et al. Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 (2004).
Elchebly, M. et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283, 1544–1548 (1999).
Kasibhatla, B., Wos, J. & Peters, K.G. Targeting protein tyrosine phosphatase to enhance insulin action for the potential treatment of diabetes. Curr. Opin. Investig. Drugs 8, 805–813 (2007).
Chiarugi, P. et al. LMW-PTP is a negative regulator of insulin-mediated mitotic and metabolic signalling. Biochem. Biophys. Res. Commun. 238, 676–682 (1997).
Bottini, N., MacMurray, J., Peters, W., Rostamkhani, M. & Comings, D.E. Association of the acid phosphatase (ACP1) gene with triglyceride levels in obese women. Mol. Genet. Metab. 77, 226–229 (2002).
Gloria-Bottini, F. et al. Phosphotyrosine protein phosphatases and diabetic pregnancy: an association between low molecular weight acid phosphatase and degree of glycemic control. Experientia 52, 340–343 (1996).
Lucarini, N. et al. Phosphotyrosine-protein-phosphatase and diabetic disorders. Further studies on the relationship between low molecular weight acid phosphatase genotype and degree of glycemic control. Dis. Markers 14, 121–125 (1998).
Iannaccone, U. et al. Serum glucose concentration and ACP1 genotype in healthy adult subjects. Metabolism 54, 891–894 (2005).
Pandey, S.K. et al. Reduction of low molecular weight protein-tyrosine phosphatase expression improves hyperglycemia and insulin sensitivity in obese mice. J. Biol. Chem. 282, 14291–14299 (2007).
Stefani, M. et al. Dephosphorylation of tyrosine phosphorylated synthetic peptides by rat liver phosphotyrosine protein phosphatase isoenzymes. FEBS Lett. 326, 131–134 (1993).
Barr, A.J. Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Med. Chem. 2, 1563–1576 (2010).
Maccari, R. & Ottanà, R. Low molecular weight phosphotyrosine protein phosphatases as emerging targets for the design of novel therapeutic agents. J. Med. Chem. 55, 2–22 (2012).
Ottanà, R. et al. 5-Arylidene-2-phenylimino-4-thiazolidinones as PTP1B and LMW-PTP inhibitors. Bioorg. Med. Chem. 17, 1928–1937 (2009).
Maccari, R. et al. Structure-based optimization of benzoic acids as inhibitors of protein tyrosine phosphatase 1B and low molecular weight protein tyrosine phosphatase. Chem. Med. Chem. 4, 957–962 (2009).
Maccari, R. et al. 5-Arylidene-2,4-thiazolidinediones as inhibitors of protein tyrosine phosphatases. Bioorg. Med. Chem. 15, 5137–5149 (2007).
Ottanà, R. et al. Synthesis, biological activity and structure-activity relationships of new benzoic acid-based protein tyrosine phosphatase inhibitors endowed with insulinomimetic effects in mouse C2C12 skeletal muscle cells. Eur. J. Med. Chem. 71, 112–127 (2014).
Forghieri, M. et al. Synthesis, activity and molecular modeling of a new series of chromones as low molecular weight protein tyrosine phosphatase inhibitors. Bioorg. Med. Chem. 17, 2658–2672 (2009).
Wade, F. et al. Deletion of low molecular weight protein tyrosine phosphatase (Acp1) protects against stress-induced cardiomyopathy. J. Pathol. 237, 482–494 (2015).
Surwit, R.S., Kuhn, C.M., Cochrane, C., McCubbin, J.A. & Feinglos, M.N. Diet-induced type II diabetes in C57BL/6J mice. Diabetes 37, 1163–1167 (1988).
McGovern, S.L., Helfand, B.T., Feng, B. & Shoichet, B.K. A specific mechanism of nonspecific inhibition. J. Med. Chem. 46, 4265–4272 (2003).
Baell, J.B. & Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Ardecky, R.J. et al. in Probe Reports from the NIH Molecular Libraries Program (National Center for Biotechnology Information, 2010).
Ramponi, G. & Stefani, M. Structure and function of the low Mr phosphotyrosine protein phosphatases. Biochim. Biophys. Acta 1341, 137–156 (1997).
Zhang, M., Zhou, M., Van Etten, R.L. & Stauffacher, C.V. Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate. Biochemistry 36, 15–23 (1997).
Rastogi, V.K. et al. 1H, 15N, and 13C resonance assignments of low molecular weight human cytoplasmic protein tyrosine phosphatase-A (HCPTP-A). J. Biomol. NMR 23, 251–252 (2002).
Su, X.D., Taddei, N., Stefani, M., Ramponi, G. & Nordlund, P. The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase. Nature 370, 575–578 (1994).
Brandão, T.A., Hengge, A.C. & Johnson, S.J. Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps. J. Biol. Chem. 285, 15874–15883 (2010).
Pannifer, A.D., Flint, A.J., Tonks, N.K. & Barford, D. Visualization of the cysteinyl-phosphate intermediate of a protein-tyrosine phosphatase by x-ray crystallography. J. Biol. Chem. 273, 10454–10462 (1998).
Kiselar, J.G., Maleknia, S.D., Sullivan, M., Downard, K.M. & Chance, M.R. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. Int. J. Radiat. Biol. 78, 101–114 (2002).
Xu, G. & Chance, M.R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 (2007).
Logan, T.M. et al. Solution structure of a low molecular weight protein tyrosine phosphatase. Biochemistry 33, 11087–11096 (1994).
Chiarugi, P. et al. LMW-PTP is a positive regulator of tumor onset and growth. Oncogene 23, 3905–3914 (2004).
Liu, P., Jenkins, N.A. & Copeland, N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13, 476–484 (2003).
Wu, S. et al. Multidentate small-molecule inhibitors of vaccinia H1-related (VHR) phosphatase decrease proliferation of cervix cancer cells. J. Med. Chem. 52, 6716–6723 (2009).
Wu, S., Bottini, M., Rickert, R.C., Mustelin, T. & Tautz, L. In silico screening for PTPN22 inhibitors: active hits from an inactive phosphatase conformation. ChemMedChem 4, 440–444 (2009).
Kholod, N. & Mustelin, T. Novel vectors for co-expression of two proteins in E. coli. Biotechniques 31, 322–323, 326–328 (2001).
Stanford, S.M. et al. Discovery of a novel series of inhibitors of lymphoid tyrosine phosphatase with activity in human T cells. J. Med. Chem. 54, 1640–1654 (2011).
Mustelin, T., Tautz, L. & Page, R. Structure of the hematopoietic tyrosine phosphatase (HePTP) catalytic domain: structure of a KIM phosphatase with phosphate bound at the active site. J. Mol. Biol. 354, 150–163 (2005).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallor. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Gupta, S., Sullivan, M., Toomey, J., Kiselar, J. & Chance, M.R. The Beamline X28C of the Center for Synchrotron Biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J. Synchrotron Radiat. 14, 233–243 (2007).
Xu, G. & Chance, M.R. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Anal. Chem. 77, 4549–4555 (2005).
Xu, G., Kiselar, J., He, Q. & Chance, M.R. Secondary reactions and strategies to improve quantitative protein footprinting. Anal. Chem. 77, 3029–3037 (2005).
Xu, H. & Freitas, M.A. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics 8, 133 (2007).
Takamoto, K. & Chance, M.R. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 (2006).
Acknowledgements
The authors are grateful to L. Tautz at the Sanford Burnham Prebys Medical Discovery Institute for providing recombinant VHR and LYP proteins, to E. Santelli for critical review of the manuscript, to the University of California Davis Mouse Biology Program for help with image preparation, to S. Gupta for assistance with sample irradiation at the Advanced Light Source of Lawrence Berkeley National Laboratory, and to Z. Mikulski and A. Lamberth for help with histological analysis at the Histology and Microscopy Core at the La Jolla Institute for Allergy and Immunology. This work was supported by grants R03DA033986 (to N.B.) and R01DK106233 from the National Institutes of Health (to N.B. and A.B.P.). S.M.S. was supported by the American Diabetes Association Pathway to Stop Diabetes Grant 1-15-INI-13 and by the University of California San Diego Diabetes Research Center grant P30DK063491 from the National Institutes of Health. The X-ray footprinting was carried out by S. Gupta at beamline 3.2.1 of the Advanced Light Source of Lawrence Berkeley National Laboratory, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. This is manuscript #1882 from the La Jolla Institute for Allergy and Immunology.
Author information
Authors and Affiliations
Contributions
S.M.S., A.E.A., T.D.Y.C., T.M., S.S., L.A.B., R.C.L., A.B.P. and N.B. participated in study conception and design. S.M.S., V.Z., M.R.B., Y.L., A.B. and S.S. performed in vivo experiments. S.M.S., V.Z. and M.R.B. performed cell biology experiments. S.M.S., V.Z., M.P.H., M.R.B., F.Y. and S.K. performed in vitro enzymatic assays. R.J.A., J.Z., S.R.G. and A.B.P. designed chemical compounds and/or performed chemical syntheses. R.J.A. and A.B.P. coordinated compound stability and PK studies. A.E.A., M.P.H., M.R.B., A.A.B., Y.L., G.W.C. and L.A.B. produced recombinant proteins. A.A.B. performed ITC experiments. A.E.A., G.W.C., J.Y. and L.A.B. performed NMR and X-ray crystallography experiments. J.K. performed hydroxyl radical footprinting experiments. S.M.S., A.E.A., V.Z., R.J.A., M.P.H., J.Z., S.R.G., M.R.B., F.Y., A.A.B., J.K., Y.L., G.W.C., S.K., J.Y., A.B., T.D.Y.C., T.M., S.S., L.A.B., R.C.L., A.B.P. and N.B. analyzed and interpreted data. S.M.S., R.C.L., A.B.P. and N.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The La Jolla Institute for Allergy and Immunology and Sanford Burnham Medical Discovery Institute hold a pending patent, WO 2016/061280 A1, “Inhibitors of low molecular weight protein tyrosine phosphatase and uses thereof,” with N.B., J.Z., S.R.G., S.M.S., A.B.P., T.D.Y.C., M.P.H. and R.J.A. named as inventors.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–19 (PDF 7192 kb)
Supplementary Note
Chemical Synthesis Information (PDF 308 kb)
Rights and permissions
About this article
Cite this article
Stanford, S., Aleshin, A., Zhang, V. et al. Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase. Nat Chem Biol 13, 624–632 (2017). https://doi.org/10.1038/nchembio.2344
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2344
This article is cited by
-
Targeting protein phosphatases in cancer immunotherapy and autoimmune disorders
Nature Reviews Drug Discovery (2023)
-
Repurposing conformational changes in ANL superfamily enzymes to rapidly generate biosensors for organic and amino acids
Nature Communications (2023)
-
Validated determination of NRG1 Ig-like domain structure by mass spectrometry coupled with computational modeling
Communications Biology (2022)
-
Differential impact of cold and hot tea extracts on tyrosine phosphatases regulating insulin receptor activity: a focus on PTP1B and LMW-PTP
European Journal of Nutrition (2022)
-
Inhibition of protein tyrosine phosphatase improves mitochondrial bioenergetics and dynamics, reduces oxidative stress, and enhances adipogenic differentiation potential in metabolically impaired progenitor stem cells
Cell Communication and Signaling (2021)