Abstract
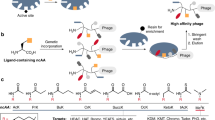
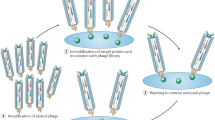
Here we describe a phage strategy for the selection of ligands based on bicyclic or linear peptides attached covalently to an organic core. We designed peptide repertoires with three reactive cysteine residues, each spaced apart by several random amino acid residues, and we fused the repertoires to the phage gene-3-protein. Conjugation with tris-(bromomethyl)benzene via the reactive cysteines generated repertoires of peptide conjugates with two peptide loops anchored to a mesitylene core. Iterative affinity selections yielded several enzyme inhibitors; after further mutagenesis and selection, we were able to chemically synthesize a lead inhibitor (PK15; Ki = 1.5 nM) specific to human plasma kallikrein that efficiently interrupted the intrinsic coagulation pathway in human plasma tested ex vivo. This approach offers a powerful means of generating and selecting bicyclic macrocycles (or if cleaved, linear derivatives thereof) as ligands poised at the interface of small-molecule drugs and biologics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bleicher, K.H., Bohm, H.J., Muller, K. & Alanine, A.I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discov. 2, 369–378 (2003).
Hüser, J. High-Throughput Screening in Drug Discovery Vol. 35 (Wiley-VCH, Weinheim, Germany, 2006).
Huse, W.D. et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 246, 1275–1281 (1989).
Ward, E.S., Gussow, D., Griffiths, A.D., Jones, P.T. & Winter, G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 341, 544–546 (1989).
McCafferty, J., Griffiths, A.D., Winter, G. & Chiswell, D.J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Scott, J.K. & Smith, G.P. Searching for peptide ligands with an epitope library. Science 249, 386–390 (1990).
Marks, J.D., Hoogenboom, H.R., Griffiths, A.D. & Winter, G. Molecular evolution of proteins on filamentous phage. Mimicking the strategy of the immune system. J. Biol. Chem. 267, 16007–16010 (1992).
Lipovsek, D. & Pluckthun, A. In-vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods 290, 51–67 (2004).
Ulrich, H. DNA and RNA aptamers as modulators of protein function. Med. Chem. 1, 199–208 (2005).
Barbas, C.F, III. Synthetic human antibodies. Nat. Med. 1, 837–839 (1995).
Holliger, P. & Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 23, 1126–1136 (2005).
Lerner, R.A. Manufacturing immunity to disease in a test tube: the magic bullet realized. Angew. Chem. Int. Edn Engl. 45, 8106–8125 (2006).
Nygren, P.A. & Skerra, A. Binding proteins from alternative scaffolds. J. Immunol. Methods 290, 3–28 (2004).
Doyon, J.B., Snyder, T.M. & Liu, D.R. Highly sensitive in vitro selections for DNA-linked synthetic small molecules with protein binding affinity and specificity. J. Am. Chem. Soc. 125, 12372–12373 (2003).
Melkko, S., Scheuermann, J., Dumelin, C.E. & Neri, D. Encoded self-assembling chemical libraries. Nat. Biotechnol. 22, 568–574 (2004).
Mannocci, L. et al. High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc. Natl. Acad. Sci. USA 105, 17670–17675 (2008).
Woiwode, T.F. et al. Synthetic compound libraries displayed on the surface of encoded bacteriophage. Chem. Biol. 10, 847–858 (2003).
Brenner, S. & Lerner, R.A. Encoded combinatorial chemistry. Proc. Natl. Acad. Sci. USA 89, 5381–5383 (1992).
Gartner, Z.J. et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science 305, 1601–1605 (2004).
Halpin, D.R. & Harbury, P.B. DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol. 2, E174 (2004).
Tse, B.N., Snyder, T.M., Shen, Y. & Liu, D.R. Translation of DNA into a library of 13,000 synthetic small-molecule macrocycles suitable for in vitro selection. J. Am. Chem. Soc. 130, 15611–15626 (2008).
Kemp, D.S. & McNamara, P.E. Conformationally restricted cyclic nonapeptides derived from L-cysteine and LL-3-amino-2-piperidino-6-carboxylic acid (LL-acp), a potent b-turn-inducing dipeptide analogue. J. Org. Chem. 50, 5834–5838 (1985).
Timmerman, P., Beld, J., Meloen, R.H. & Puijk, W.C. Method for selecting a candidate drug compound. WO patent 2004077062 (2004).
Timmerman, P., Beld, J., Puijk, W.C. & Meloen, R.H. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. ChemBioChem 6, 821–824 (2005).
Timmerman, P., Puijk, W.C., Slootstra, J.W., Van Dijk, E. & Meloen, R.H. Binding compounds, immunogenic compounds and peptidomimetics. WO patent 2006078161 (2006).
Jespers, L.S.A., Winter, G.P., Bonnert, T.P. & Simon, T.M. SBP members with a chemical moiety covalently bound within the binding site. WO patent 9501438 (1995).
Jespers, L., Bonnert, T.P. & Winter, G. Selection of optical biosensors from chemisynthetic antibody libraries. Protein Eng. Des. Sel. 17, 709–713 (2004).
Wessjohann, L.A., Ruijter, E., Garcia-Rivera, D. & Brandt, W. What can a chemist learn from nature's macrocycles?–a brief, conceptual view. Mol. Divers. 9, 171–186 (2005).
Driggers, E.M., Hale, S.P., Lee, J. & Terrett, N.K. The exploration of macrocycles for drug discovery–an underexploited structural class. Nat. Rev. Drug Discov. 7, 608–624 (2008).
Kather, I., Bippes, C.A. & Schmid, F.X. A stable disulfide-free gene-3-protein of phage fd generated by in vitro evolution. J. Mol. Biol. 354, 666–678 (2005).
Cheng, Y. & Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973).
Abbenante, G. & Fairlie, D.P. Protease inhibitors in the clinic. Med. Chem. 1, 71–104 (2005).
Turk, B. Targeting proteases: successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799 (2006).
Millward, S.W., Fiacco, S., Austin, R.J. & Roberts, R.W. Design of cyclic peptides that bind protein surfaces with antibody-like affinity. ACS Chem. Biol. 2, 625–634 (2007).
Litovchick, A. & Szostak, J.W. Selection of cyclic peptide aptamers to HCV IRES RNA using mRNA display. Proc. Natl. Acad. Sci. USA 105, 15293–15298 (2008).
Cremlyn, R.J. An Introduction to Organosulfur Chemistry 1st edn. (Wiley, Chichester, UK, 1996).
Wood, S.P. et al. Crystal structure analysis of deamino-oxytocin: conformational flexibility and receptor binding. Science 232, 633–636 (1986).
Bhaskaran, R., Chuang, L.C. & Yu, C. Conformational properties of oxytocin in dimethyl sulfoxide solution: NMR and restrained molecular dynamics studies. Biopolymers 32, 1599–1608 (1992).
Pohl, E. et al. Structure of octreotide, a somatostatin analogue. Acta Crystallogr. D Biol. Crystallogr. 51, 48–59 (1995).
Melacini, G., Zhu, Q. & Goodman, M. Multiconformational NMR analysis of sandostatin (octreotide): equilibrium between beta-sheet and partially helical structures. Biochemistry 36, 1233–1241 (1997).
Jackson, D.Y. et al. A designed peptide ligase for total synthesis of ribonuclease A with unnatural catalytic residues. Science 266, 243–247 (1994).
Sandman, K.E. & Noren, C.J. The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 28, 755–761 (2000).
Sandman, K.E., Benner, J.S. & Noren, C.J. Phage display of selenopeptides. J. Am. Chem. Soc. 122, 960–961 (2000).
Dennis, M.S. et al. Peptide exosite inhibitors of factor VIIa as anticoagulants. Nature 404, 465–470 (2000).
Huang, L. et al. Novel peptide inhibitors of angiotensin-converting enzyme 2. J. Biol. Chem. 278, 15532–15540 (2003).
Acknowledgements
We thank P. Jones (Laboratory of Molecular Biology, Cambridge, UK) for expert laboratory advice, L. Judd (Centre for Protein Engineering, Cambridge, UK) for media preparation, A. Jaulent (Centre for Protein Engineering, Cambridge, UK) for peptide purification, and F. Begum (Laboratory of Molecular Biology, Cambridge, UK) and S.-Y. Peak-Chew (Laboratory of Molecular Biology, Cambridge, UK) for mass spectrometric analysis. We also thank I. Kather and F.X. Schmid from the University of Bayreuth for the engineered phage with disulfide-free gene-3-protein. C.H. was supported by the Swiss National Science Foundation (SNSF) and the Novartis Foundation (formerly Ciba-Geigy Jubilee Foundation).
Author information
Authors and Affiliations
Contributions
C.H. and G.W. conceived the experiments, analyzed the data and wrote the article; C.H. performed the experiments; T.R. and S.F. solved the NMR structure.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1 and 2, and Supplementary Methods (PDF 325 kb)
Rights and permissions
About this article
Cite this article
Heinis, C., Rutherford, T., Freund, S. et al. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat Chem Biol 5, 502–507 (2009). https://doi.org/10.1038/nchembio.184
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.184
This article is cited by
-
Selenium chemistry for spatio-selective peptide and protein functionalization
Nature Reviews Chemistry (2024)
-
Development of cyclic peptides that can be administered orally to inhibit disease targets
Nature Chemical Biology (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)
-
Extendable stapling of unprotected peptides by crosslinking two amines with o-phthalaldehyde
Nature Communications (2022)