Abstract
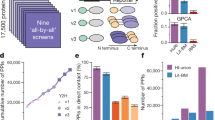
ATP-binding cassette (ABC) transporters are a ubiquitous class of integral membrane proteins of immense clinical interest because of their strong association with human disease and pharmacology. To improve our understanding of these proteins, we used membrane yeast two-hybrid technology to map the protein interactome of all of the nonmitochondrial ABC transporters in the model organism Saccharomyces cerevisiae and combined this data with previously reported yeast ABC transporter interactions in the BioGRID database to generate a comprehensive, integrated 'interactome'. We show that ABC transporters physically associate with proteins involved in an unexpectedly diverse range of functions. We specifically examine the importance of the physical interactions of ABC transporters in both the regulation of one another and in the modulation of proteins involved in zinc homeostasis. The interaction network presented here will be a powerful resource for increasing our fundamental understanding of the cellular role and regulation of ABC transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lewis, V.G., Ween, M.P. & McDevitt, C.A. The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 249, 919–942 (2012).
Verrier, P.J. et al. Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159 (2008).
Dean, M., Rzhetsky, A. & Allikmets, R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166 (2001).
Dean, M. & Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6, 123–142 (2005).
Paumi, C.M., Chuk, M., Snider, J., Stagljar, I. & Michaelis, S. ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593 (2009).
Hopfner, K.P. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13, 249–255 (2003).
Jones, P.M., O'Mara, M.L. & George, A.M. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531 (2009).
Jones, P.M. & George, A.M. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol. Life Sci. 61, 682–699 (2004).
Gottesman, M.M. & Ambudkar, S.V. Overview: ABC transporters and human disease. J. Bioenerg. Biomembr. 33, 453–458 (2001).
Cannon, R.D. et al. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22, 291–321 (2009).
Szakács, G., Paterson, J.K., Ludwig, J.A., Booth-Genthe, C. & Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5, 219–234 (2006).
Stagljar, I., Korostensky, C., Johnsson, N. & Te Heesen, S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. USA 95, 5187–5192 (1998).
Snider, J. et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat. Protoc. 5, 1281–1293 (2010).
Kovalchuk, A. & Driessen, A.J.M. Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11, 177 (2010).
Mason, D.L., Mallampalli, M.P., Huyer, G. & Michaelis, S. A region within a lumenal loop of Saccharomyces cerevisiae Ycf1p directs proteolytic processing and substrate specificity. Eukaryot. Cell 2, 588–598 (2003).
Paumi, C.M. et al. Mapping protein-protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid analysis. Mol. Cell 26, 15–25 (2007).
Kelm, K.B., Huyer, G., Huang, J.C. & Michaelis, S. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic 5, 165–180 (2004).
Kölling, R. & Hollenberg, C.P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13, 3261–3271 (1994).
Hu, C.D., Chinenov, Y. & Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Gelperin, D.M. et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005).
Stark, C. et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 39, D698–D704 (2011).
Cherry, J.M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 (2012).
Shani, N. & Valle, D. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl. Acad. Sci. USA 93, 11901–11906 (1996).
Kolaczkowska, A., Kolaczkowski, M., Goffeau, A. & Moye-Rowley, W.S. Compensatory activation of the multidrug transporters Pdr5p, Snq2p, and Yor1p by Pdr1p in Saccharomyces cerevisiae. FEBS Lett. 582, 977–983 (2008).
Giaever, G. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002).
Rogers, B. et al. The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 3, 207–214 (2001).
MacDiarmid, C.W., Milanick, M.A. & Eide, D.J. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277, 39187–39194 (2002).
Zhao, H. & Eide, D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93, 2454–2458 (1996).
Gitan, R.S., Shababi, M., Kramer, M. & Eide, D.J. A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J. Biol. Chem. 278, 39558–39564 (2003).
MacDiarmid, C.W., Milanick, M.A. & Eide, D.J. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278, 15065–15072 (2003).
Rockwell, N.C., Wolfger, H., Kuchler, K. & Thorner, J. ABC transporter Pdr10 regulates the membrane microenvironment of Pdr12 in Saccharomyces cerevisiae. J. Membr. Biol. 229, 27–52 (2009).
Cabrito, T.R., Teixeira, M.C., Singh, A., Prasad, R. & Sá-Correia, I. The yeast ABC transporter Pdr18 (ORF YNR070w) controls plasma membrane sterol composition, playing a role in multidrug resistance. Biochem. J. 440, 195–202 (2011).
Pomorski, T. et al. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14, 1240–1254 (2003).
Li, Y. & Prinz, W.A. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 279, 45226–45234 (2004).
Wilcox, L.J. et al. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277, 32466–32472 (2002).
Eide, D.J. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284, 18565–18569 (2009).
Burke, M.A., Mutharasan, R.K. & Ardehali, H. The sulfonylurea receptor, an atypical ATP-binding cassette protein, and its regulation of the KATP channel. Circ. Res. 102, 164–176 (2008).
Degryse, E., Dumas, B., Dietrich, M., Laruelle, L. & Achstetter, T. In vivo cloning by homologous recombination in yeast using a two-plasmid–based system. Yeast 11, 629–640 (1995).
Sung, M.K. & Huh, W. Bimolecular fluorescence complementation analysis system for in vivo detection of protein–protein interaction in Saccharomyces cerevisiae. Yeast 24, 767–775 (2007).
Longtine, M.S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Lee, M.E. et al. The Rho1 GTPase acts together with a vacuolar glutathione S-conjugate transporter to protect yeast cells from oxidative stress. Genetics 188, 859–870 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Boyle, E.I. et al. GO:TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715 (2004).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Ostlund, G. et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38, D196–D203 (2010).
Amberger, J., Bocchini, C.A., Scott, A.F. & Hamosh, A. McKusick's Online Mendelian Inheritance in Man (OMIM). Nucleic Acids Res. 37, D793–D796 (2009).
Babu, M., Krogan, N.J., Awrey, D.E., Emili, A. & Greenblatt, J.F. Systematic characterization of the protein interaction network and protein complexes in Saccharomyces cerevisiae using tandem affinity purification and mass spectrometry. Methods Mol. Biol. 548, 187–207 (2009).
Acknowledgements
This work is dedicated to our late friend and colleague Igor Shevelev. We thank B. Andrews, C. Nislow, F. Vizeacoumar, C. Kurat and S. Alfred (University of Toronto) for providing reagents, assistance and equipment; L. Miller, S. Pagant, K. Kuchler and C. Klein for experimental assistance; and W.K. Huh (Seoul National University) for providing BiFC plasmids. We also thank V. Kanelis, K. Sokolina and M. Ali for reviewing the manuscript. This work was supported by grants from the Canadian Institutes of Health Research, Canadian Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, Ontario Genomics Institute, Canadian Cystic Fibrosis Foundation, Canadian Cancer Society, University Health Network to I. Stagljar and National Institutes of Health (R01-GM76375 to H.-O.P. and R01-GM51508 to S.M.).
Author information
Authors and Affiliations
Contributions
I. Stagljar designed the project. J.S. was actively involved in all experiments, and wrote the bulk of the manuscript. I. Stagljar and S.M. provided project guidance and assisted in manuscript preparation. K.J. and Z.Z. performed bioinformatics analysis. H.-O.P. and M.E.L. performed BiFC experiments. A.R.Y. performed Westerns, phenotype assays and strain generation. A.H. carried out MYTH screening and zinc-related functional analysis. M.C., D.D., C.G., M.W., P.T. and V.W. carried out MYTH screening. S.L.S. and D.B. carried out MYTH screening of AUS1 under anaerobic conditions. K.D.D. carried out the PDR11 and AUS1 western blots. C.G., M.J., J.F.G and M.B. performed co-IP experiments. C.B. and B.-J.S.L. provided deletion strains. C.M.P. carried out zinc transport assays. H.-O.P. and C.M.P. critically reviewed the manuscript. I. Shevelev was involved in bait strain generation and initial project design.
Corresponding author
Ethics declarations
Competing interests
I. Stagljar is cofounder of Dualsystems Biotech, Switzerland.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–25 and Supplementary Tables 1–2 (PDF 5999 kb)
Supplementary Data Set 1
Comprehensive list of all hits identified in MYTH screening. (XLSX 16 kb)
Supplementary Data Set 2
Comprehensive list of all members of the integrated ABC transporter interactome annotated to identify those with human orthologs and human orthologs associated with (XLSX 34 kb)
Rights and permissions
About this article
Cite this article
Snider, J., Hanif, A., Lee, M. et al. Mapping the functional yeast ABC transporter interactome. Nat Chem Biol 9, 565–572 (2013). https://doi.org/10.1038/nchembio.1293
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1293
This article is cited by
-
Prognostic value and immune infiltration of novel signatures in colon cancer microenvironment
Cancer Cell International (2021)
-
Engineering endogenous ABC transporter with improving ATP supply and membrane flexibility enhances the secretion of β-carotene in Saccharomyces cerevisiae
Biotechnology for Biofuels (2020)
-
A drug discovery platform to identify compounds that inhibit EGFR triple mutants
Nature Chemical Biology (2020)
-
Transport of Anthocyanins and other Flavonoids by the Arabidopsis ATP-Binding Cassette Transporter AtABCC2
Scientific Reports (2019)
-
Development of Series of Affinity Tags in Streptomyces
Scientific Reports (2017)