Abstract
Apicobasal polarity is known to affect epithelial morphogenesis and cell differentiation, but it remains unknown how these processes are mechanistically orchestrated. We find that ligand-specific EGFR signalling via PI(3)K and Rac1 autonomously modulates apicobasal polarity to enforce the sequential control of morphogenesis and cell differentiation. Initially, EGF controls pancreatic tubulogenesis by negatively regulating apical polarity induction. Subsequently, betacellulin, working via inhibition of atypical protein kinase C (aPKC), causes apical domain constriction within neurogenin3+ endocrine progenitors, which results in reduced Notch signalling, increased neurogenin3 expression, and β-cell differentiation. Notably, the ligand-specific EGFR output is not driven at the ligand level, but seems to have evolved in response to stage-specific epithelial influences. The EGFR-mediated control of β-cell differentiation via apical polarity is also conserved in human neurogenin3+ cells. We provide insight into how ligand-specific EGFR signalling coordinates epithelial morphogenesis and cell differentiation via apical polarity dynamics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
13 November 2017
In the version of this Article originally published, an incorrect file was used for Supplementary Figure 4. This file has now been replaced with the correct Supplementary Figure in the online version of the Article.
References
Kesavan, G. et al. Cdc42-mediated tubulogenesis controls cell specification. Cell 139, 791–801 (2009).
Szymaniak, A. D., Mahoney, J. E., Cardoso, W. V. & Varelas, X. Crumbs3-mediated polarity directs airway epithelial cell fate through the hippo pathway effector yap. Dev. Cell 34, 283–296 (2015).
Villasenor, A., Chong, D. C., Henkemeyer, M. & Cleaver, O. Epithelial dynamics of pancreatic branching morphogenesis. Development 137, 4295–4305 (2010).
Clark, B. S. et al. Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis. Development 139, 1599–1610 (2012).
Ossipova, O., Ezan, J. & Sokol, S. Y. PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Dev. Cell 17, 222–233 (2009).
Margolis, B. & Borg, J. P. Apicobasal polarity complexes. J. Cell Sci. 118, 5157–5159 (2005).
Assemat, E., Bazellieres, E., Pallesi-Pocachard, E., Le Bivic, A. & Massey-Harroche, D. Polarity complex proteins. Biochim. Biophys. Acta 1778, 614–630 (2008).
Roignot, J., Peng, X. & Mostov, K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb. Perspect. Biol. 5, a013789 (2013).
Arntfield, M. E. & van der Kooy, D. β-Cell evolution: how the pancreas borrowed from the brain: the shared toolbox of genes expressed by neural and pancreatic endocrine cells may reflect their evolutionary relationship. Bioessays 33, 582–587 (2011).
Wang, S. et al. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev. Biol. 339, 26–37 (2010).
Bechard, M. E. et al. Precommitment low-level Neurog3 expression defines a long-lived mitotic endocrine-biased progenitor pool that drives production of endocrine-committed cells. Genes Dev. 30, 1852–1865 (2016).
Bankaitis, E. D., Bechard, M. E. & Wright, C. V. Feedback control of growth, differentiation, and morphogenesis of pancreatic endocrine progenitors in an epithelial plexus niche. Genes Dev. 29, 2203–2216 (2015).
Chartier, F. J., Hardy, E. J. & Laprise, P. Crumbs controls epithelial integrity by inhibiting Rac1 and PI3K. J. Cell Sci. 124, 3393–3398 (2011).
Miettinen, P. J. et al. Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. Development 127, 2617–2627 (2000).
Duvillie, B. et al. The mesenchyme controls the timing of pancreatic β-cell differentiation. Diabetes 55, 582–589 (2006).
Gouzi, M., Kim, Y. H., Katsumoto, K., Johansson, K. & Grapin-Botton, A. Neurogenin3 initiates stepwise delamination of differentiating endocrine cells during pancreas development. Dev. Dyn. 240, 589–604 (2011).
Kesavan, G. et al. Cdc42/N-WASP signaling links actin dynamics to pancreatic beta cell delamination and differentiation. Development 141, 685–696 (2014).
Huotari, M. A. et al. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology 143, 4437–4446 (2002).
Shing, Y. et al. Betacellulin: a mitogen from pancreatic beta cell tumors. Science 259, 1604–1607 (1993).
Guo, T., Landsman, L., Li, N. & Hebrok, M. Factors expressed by murine embryonic pancreatic mesenchyme enhance generation of insulin-producing cells from hESCs. Diabetes 62, 1581–1592 (2013).
Miyagawa, J. et al. Immunohistochemical localization of betacellulin, a new member of the EGF family, in normal human pancreas and islet tumor cells. Endocr. J. 46, 755–764 (1999).
Benitez, C. M. et al. An integrated cell purification and genomics strategy reveals multiple regulators of pancreas development. PLoS Genet. 10, e1004645 (2014).
Ekberg, J., Hansson, M., Døhn, U., Hess, K. & Funa, N. Generation of pancreatic endoderm from pluripotent stem cells using small molecules. Patent WO 2014/033322 A1 (2014).
Døhn, U., Christophersen, N. S. & Ekberg, J. Generation of endocrine progenitor cells from human pluripotent stem cells using small molecules. Patent WO 2015/028614 (2015).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Del Bene, F., Wehman, A. M., Link, B. A. & Baier, H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell 134, 1055–1065 (2008).
Ohata, S. et al. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron 69, 215–230 (2011).
Castanieto, A., Johnston, M. J. & Nystul, T. G. EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. eLife 3, e04437 (2014).
Wilson, K. J., Gilmore, J. L., Foley, J., Lemmon, M. A. & Riese, D. J. II Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol. Ther. 122, 1–8 (2009).
Foerster, S. et al. Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. Proteomics 13, 3131–3144 (2013).
Glogauer, M. et al. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J. Immunol. 170, 5652–5657 (2003).
Schonhoff, S. E., Giel-Moloney, M. & Leiter, A. B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 270, 443–454 (2004).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Zhao, J. J. et al. The p110α isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc. Natl Acad. Sci. USA 103, 16296–16300 (2006).
Threadgill, D. W. et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269, 230–234 (1995).
Mellitzer, G. et al. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Mol. Endocrinol. 18, 2765–2776 (2004).
Warming, S., Costantino, N., Court, D. L., Jenkins, N. A. & Copeland, N. G. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005).
Ahnfelt-Ronne, J. et al. An improved method for three-dimensional reconstruction of protein expression patterns in intact mouse and chicken embryos and organs. J. Histochem. Cytochem. 55, 925–930 (2007).
Potter, L. A., Choi, E., Hipkens, S. B., Wright, C. V. & Magnuson, M. A. A recombinase-mediated cassette exchange-derived cyan fluorescent protein reporter allele for Pdx1. Genesis 50, 384–392 (2012).
Sugiyama, T., Rodriguez, R. T., McLean, G. W. & Kim, S. K. Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc. Natl Acad. Sci. USA 104, 175–180 (2007).
Edsbagge, J. et al. Vascular function and sphingosine-1-phosphate regulate development of the dorsal pancreatic mesenchyme. Development 132, 1085–1092 (2005).
Percival, A. C. & Slack, J. M. Analysis of pancreatic development using a cell lineage label. Exp. Cell Res. 247, 123–132 (1999).
Fischer, Y. et al. NANOG reporter cell lines generated by gene targeting in human embryonic stem cells. PLoS ONE 5, e12533 (2010).
D’Haene, B., Vandesompele, J. & Hellemans, J. Accurate and objective copy number profiling using real-time quantitative PCR. Methods 50, 262–270 (2010).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Funa, N. S. et al. β-Catenin regulates primitive streak induction through collaborative interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 16, 639–652 (2015).
Acknowledgements
We thank K. Schachter for careful reading and editing of the manuscript and help with western blots, A. Lundqvist, J. Larsen, D. Klüver Hansen, K. Stohlmann and A. Stiehm for technical assistance, and G. Karemore, S. Heilmann and A. L. Jackson for help with statistical analysis. We are grateful to the DanStem FCCF and the FACS Core at Lund SCC for FACS assistance, CFIM for use of microscopes and the transgenic core facility at the University of Copenhagen for oocyte injections. We thank Novo Nordisk A/S for access to their proprietary pancreatic endoderm and endocrine progenitor (PE and EP) differentiation protocols as well as for sponsoring the establishment of the human NGN3-reporter line. We thank X. Varelas at Boston University School of Medicine for the Crb3 antibody. This work was supported by the Swedish Foundation for Strategic Research, the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) as part of the Beta Cell Biology Consortium (1 UO1 DK089570-01), the Juvenile Diabetes Research Foundation, the Novo Nordisk Foundation, the Danish Strategic Research Council, the Lundbeck Foundation (R100-A9422) and the Danish Council for Independent Research (ID: DFF—1331-00310A).
Author information
Authors and Affiliations
Contributions
Z.M.L.-Ö. designed, conducted and analysed experiments in all figures, made the models and wrote the paper. P.N. assisted with experimental and statistical analysis, made and characterized the Muc1mCherry reporter mouse, designed and helped conduct the live imaging, and edited the paper. M.E.B., E.B. and C.V.W. made the Ngn3RG1 reporter mouse and edited the paper. K.H. generated and validated the NGN3GFP hESC reporter. T.U.G. designed and conducted the experiments in Fig. 5 and Supplementary Fig. 2. J.A. assisted with cell culturing, performed the siRNA transfections, designed qPCR primers and edited the paper. H.S. designed experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Reporter constructs and apicobasal polarity in Ngn3 cells at E15.5 and E12.5.
(a) Construct of the Ngn3RG1 reporter. (b) Representative section of an E15.5 Ngn3RG1 pancreas, stained for Neurogenin3 (Ngn3; white) and E-cadherin (E-cad; blue) shows that the mCherry (reporter expression; red) and GFP (reporter expression; green) overlaps with Ngn3 in the nucleus and E-cad at the membrane of the cells. (c) Construct of the Muc1mCherry reporter. (d) Representative section of a Muc1mCherry positive pancreas from an adult mouse, stained for Mucin1 (Muc1; green) shows that the mCherry (reporter expression; red) overlaps with Muc1 at the apical membrane. (e) Representative section of E15.5 WT pancreata (6 μm) stained for neurogenin3 (Ngn3; red), atypical protein kinase C (aPKC;green), mucin1 (Muc1; blue) and E-cadherin (Ecad; white). (f) Representative section of E15.5 WT pancreata (6 μm) stained for neurogenin3 (Ngn3; red), Crumbs3 (Crb3;green), mucin1 (Muc1; blue) and E-cadherin (Ecad; white). (g) Representative flow cytometry data from E12.5 Ngn3RG1 pancreata stained for Muc1, n = 3. (h) Quantification of the fraction of Ngn3 + cells that are Muc1 + (polarized) or Muc1- (Unpolarized) at E12.5, analyzed by Flow cytometry. n = 3 pancreata from each condition, from two litters, pTT < 0.0001. Error bars represent ± SEM. Scale bars, 20 μm. Statistical analysis: two-tailed student’s t-test. Statistic source data are found in Supplementary Table 3.
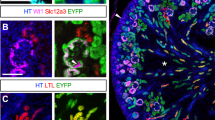
Supplementary Figure 2 Rac1 regulates apical polarity induction in multipotent progenitors and apical domain size in Ngn3 cells.
(a) Representative Rac1 western blot in E15.5 Ctrl and Rac1;pdxCre KO (Loading control: α-tubulin). (b) Representative 3D-projections of E14.5 Ctrl and Rac1;pdxCre KO dorsal pancreata stained for Muc1. (c) Polarity quantification in E15.5 Ctrl and Rac1;pdxCre KO. n = 3 pancreata, from two litters, pTT = 0.0324. (d,e) Representative sections of E12.5 Ctrl and Rac1;pdxCreER KO, stained for β-gal, Muc1 and E-cad, and polarity quantification. n = 3 pancreata, from two litters, pTT = 0.0002. (f,g) Apical width quantification of E12.5 and E15.5 Ctrl and WT/KO cells in Rac1;pdxCreER KO. E12.5: n = 3 pancreata (from two litters), Het/KO pTT = 0.356 (n.s.), Het/WT pTT = 0.942 (n.s.). E15.5: n = 3 pancreata (from two litters), pTT = 0.674 (n.s.. h) Representative sections of E15.5 Ctrl and Rac1;pdxCre KO stained for Ngn3 and Ecad. (i–k) Ngn3/Ecad mm2 (i), Sox9/Ecad mm2 (j) and Ngn3 + /Sox9 + quantification in E15.5 Ctrl and Rac1;pdxCre KO. (i) n = 4 pancreata (two litters), pTT = 0.0394; (j) n = 3 pancreata, from two litters, pTT = 0.0122; (k) n = 4 pancreata, from two litters, pTT = 0.7289. (l) Representative sections of E15.5 Ctrl and Rac1;pdxCre KO stained for Ngn3 and Ecad. (m) qPCR analysis of Ngn3 in E15.5 Ctrl and Rac1;pdxCre KO (normalization to β-actin). n = 7 Ctrl, 13 KO pancreata, (three litters), pTT = 0.0022. (n) Ngn3 intensity quantification in E15.5 Ctrl and Rac1;pdxCre KO. n = 3 pancreata from each condition (two litters), pTT = 0.01011. (o,p) Representative flow cytometry plots and quantification of%Ngn3High cells in E15.5 Ctrl and Rac1;pdxCre KO. n = 4 Ctrl, 5 KO pancreata, from two independent experiments, pTT = 0.0093. (q) Apical width quantification from E15.5 Ctrl and Rac1;pdxCre KO. n = 3 pancreata (two litters), pTT = 0.0132. (r) Apical volume quantification from E15.5 Ctrl and Rac1;pdxCre KO. n = 3 pancreata (two litters), pKW = 0.04953. (s,t) Representative sections of E17.5 Ctrl and Rac1;pdxCre KO stained for Insulin (Ins) and Ecad and quantification of β- and α-cell differentiation. n = 4 pancreata (two litters), Insulin(β): pTT = 0.0326, Glucagon(α): pTT = 0.6323. Error bars represent ± SEM. Scale bars, 20 μm (d,l), 50 μm (b,h), 100 μm (r). Statistic source data are found in Supplementary Table 3. Unprocessed blots are shown in Supplementary Fig. 11. Statistical analysis: two-tailed student’s t-test (pTT) or Kruskal-Wallis (pKW).
Supplementary Figure 3 The PI3K/Rac1 feedback loop promotes protection from ubiquitination-mediated proteasome degradation.
(a) Relative band intensity for Rac1 western blots in Fig. 3a, in E11.5 + 1 Ctrl or LY-treated explants. n = 3 explants (two litters), pTT = 0.0008. (b) Relative band intensity for p85 western blots in Fig. 3b, in E11.5 Ctrl and Rac1;pdxCre KO pancreata. n = 3 pancreata (two litters), pTT = 0.0041. (c,d) Representative western blots and relative band intensity from E11.5 Ctrl and LY-treated explants treated with or without the proteasome inhibitor (MG) (6h). n = 3 explants, from two independent experiments, Ctrl/LY pTT < 0.0001, Ctrl/LY + MG pTT = 0.1296. (e) Representative sections of E15.5 pancreata and E12.5 + 3 explants, stained for Ngn3, Ecad and DAPI. (f) Ngn3/Ecad mm2 quantification in E15.5 pancreata and E12.5 + 3 explants. n = 5 pancreata or explants (two litters each), pTT = 0.9282. (g) Mean Ngn3 intensity quantification (Ngn3+ cells) in E15.5 pancreata and E12.5 + 3 explants. n = 3 pancreata or explants, (two litters each), pTT = 0.2948 (n.s.). (h,i) Representative flow Cytometry plots and quantification of %Ngn3High cells from Ngn3Y FP+ E15.5 pancreata and E12.5 + 3 explants. n = 6 pancreata or explants, (two litters each), pTT = 0.5005. (j) β-cell differentiation (Insulin +) quantification in E17.5 pancreata and E11.5 + 7 explants. n = 5 pancreata or explants (two litters each), pTT = 0.6608. (k) Representative sections of E17.5 pancreata and E11.5 + 7 explants stained for Insulin, Ecad and DAPI. (l) Representative sections of E11.5 + 3 Ctrl and LY-treated explants stained for PHH3, Ecad and DAPI. (m,n) Proliferation quantification in E11.5 + 3 Ctrl and LY-treated explants (m) or E15.5 Ctrl and Rac1;pdxCre KO pancreata (n). (m) n = 3 pancreatic explants (two litters), pTT = 0.0413; (n) n = 3 pancreata, (two litters), pTT = 0.8791. (o) Apoptosis quantification in E11.5 + 3 Ctrl and LY-treated explants. n = 3 pancreatic explants, (two litters), pTT = 0.5832. (p) Manual quantification of Ngn3Low to Ngn3High conversion time (minutes) from real-time imaging experiments (Supplementary Videos 2 and 3) on E12.5 + 3 Ctrl and LY-treated Ngn3RG1/Muc1mCherry pancreatic explants. n = 6 Ctrl, 5 LY cells from one experiment, pTT = 0.0025. Error bars represent ± SEM. Scale bars, 25 μm (e), 50 μm (k,l). Statistic source data are found in Supplementary Table 3. Unprocessed blots are shown in Supplementary Fig. 11. Statistical analysis: two-tailed student’s t-test (pTT).
Supplementary Figure 4 Inhibition of PI3K or EGFR-signaling increase lumen formation and decreases β-cell differentiation.
(a) Representative 3D-projections of E11.5 + 7 Ctrl, LY- or EGFRi-treated explants, stained for E-cad (Outlined;Not shown), Muc1 and Insulin (Ins). (b) Luminal volume in E11.5 + 7 Ctrl and LY-treated explants. n = 4 explants (two litters), p = 0.0084. (c,d) Ngn3/Ecad mm2 quantification (c), and mean apical volume quantification (Ngn3+ cells) (d) in E11.5 + 3 Ctrl and LY-treated explants. (c) n = 3 explants from each condition (two litters), p = 0.0101; (d) n = 3 explants (Total number of cells: 34 Ctrl, 39 LY), pTT = 0.0076. (e) β-cell differentiation (Volume Insulin) quantification in E11.5 + 7 Ctrl and LY-treated explants. n = 3 Ctrl, 4 LY explants (two litters), pTT = 0.0004. (f,g) Representative flow cytometry plots and quantification of %Ngn3High cells in E11.5 + 3 Ngn3Y FP+ Ctrl, LY- or EGFRi-treated explants. n = 6 Ctrl, 5 LY, 3 EGFRi explants, from two independent experiments, Ctrl/LY pTT = 0.0018, Ctrl/EGFRi pTT = 0.0131. (h) Representative sections of E11.5 + 7 Ctrl, LY- or EGFRi-treated explants, stained for E-cad, Glucagon (Gluc) and DAPI. (i,j) α-cell (Glucagon +) differentiation quantifications in E11.5 + 7 Ctrl, LY- (i) or EGFRi-treated (j) explants. LY: n = 3 explants, from two litters, pTT = 0.7691; EGFRi: n = 3 explants (two litters), pTT = 0.6728. (k) Luminal Volume quantification in E11.5 + 7 Ctrl and EGFRi-treated explants. n = 7 Ctrl, 5 EGFRi explants, from three litters, pTT = 0.0351. (l) Ngn3/Ecad mm2 quantification in E11.5 + 3 Ctrl and EGFRi-treated explants. n = 5 explants, from three litters, pTT = 0.0224. (m) β-cell differentiation (Volume Insulin) quantification in E11.5 + 7 Ctrl and EGFRi-treated explants. n = 5 Ctrl, 8 EGFRi explants, from three litters. pTT = 0.0073. (n,o) Relative band intensity quantifications of Rac1 (n) and p85 (o) on western blots in Fig. 3i. Rac1: n = 5 protein samples, from two litters, pTT = 0.0125; p85: n = 4 Ctrl, 3 EGFRi protein samples, from two litters, pTT = 0.0438. (p) Representative flow cytometry plots and quantification of %Ngn3High cells in E12.5 + 3 Ngn3Y FP+ Ctrl and aPKCi-treated explants. n = 4 explants (two litters), pTT = 0.0059. (r) Representative 3D-projections of E12.5 + 7 Ctrl, aPKCi- or CpE-treated (from E12.5 + 4 and onwards) explants stained for E-cad (Outlined;Not shown), Muc1 and Insulin (Ins). Error bars represent ± SEM. Scale Bars, 200 μm (a), 40 μm (h), 100 μm (p). Statistic source data are found in Supplementary Table 3. Statistical analysis: two-tailed student’s t-test (pTT).
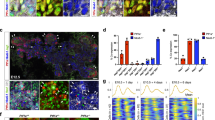
Supplementary Figure 5 EGF and BTC activate EGFR sequentially.
(a) Representative section of E11.5 WT pancreata (6 μm) stained for mucin1 (Muc1;green), EGF (red), E-cadherin (Outlined;Not shown) and DAPI (blue). (b–b ′) Representative section of E15.5 WT pancreata (6 μm) stained for Neurogenin3 (Ngn3;green), mucin1 (Muc1;blue), EGF (red) and E-cadherin (E-cad; White (b); Outlined;Not shown (b ′)). Indicated region visualizes area of inset in b ′. (c) Representative section of E11.5 WT pancreata (6 μm) stained for mucin1 (Muc1;green), Betacellulin (BTC;red), E-cadherin (Outlined; Not shown) and DAPI (blue). (d–d ′) Representative section of E15.5 WT pancreata (6 μm) stained for Neurogenin3 (Ngn3;green), mucin1 (Muc1;blue), Betacellulin (BTC;red) and E-cadherin (E-cad; White (d); Outlined;Not shown (d ′)). Indicated region visualizes area of inset in d ′. (e) Representative section of E11.5 WT pancreata (6 μm) stained for mucin1 (Muc1;green), EGFR (red), E-cadherin (Outlined;Not shown) and DAPI (blue). (f–f ′) Representative section of E15.5 WT pancreata (6 μm) stained for Neurogenin3 (Ngn3;green), mucin1 (Muc1;blue), EGFR (red) and E-cadherin (E-cad; White (f); Outlined;Not shown (f ′)). Indicated region visualizes area of inset in f ′. (g) Representative section of E11.5 WT pancreata (6 μm) stained for mucin1 (Muc1;green), ErbB4 (red), E-cadherin (Outlined;Not shown) and DAPI (blue). (h–h ′) Representative section of E15.5 WT pancreata (6 μm) stained for Neurogenin3 (Ngn3;green), mucin1 (Muc1;blue), ErbB4 (red) and E-cadherin (E-cad; White (h); Outlined;Not shown (h ′)). Indicated region visualizes area of inset in h ′. (i) Representative western blot analysis of E11.5 or E12.5 explants treated with EGF or BTC (0, 10, 20 or 30 ng/ml) for 48 h, when added at E11.5 or E12.5 + 2, were analyzed for activation of EGFR (p-EGFR) during the primary or secondary transition. (Loading control: Vinculin). (j) Relative band intensity (p-EGFR/Vinculin) quantification from western blots in i. n = 3 protein samples, from two independent experiments, primary transition: EGF: 0/10 pTT = 0.1379, 0/20 pTT = 0.0029, 0/30 pTT = 0.2421; BTC: 0/10 pTT = 0.5939, 0/20 pTT = 0.1592, 0/30 pTT = 0.1247; secondary transition: EGF: 0/10 pTT = 0.7233, 0/20 pTT = 0.3370, 0/30 pTT = 0.2270; BTC: 0/10 pTT = 0.1050, 0/20 pTT = 0.0001, 0/30 pTT = 0.0017. Error bars represent ± SEM. Scale bar, 40 μm (a,c,e,g), 20 μm (b,b ′,d,d ′,f,f ′,h,h ′). Statistical analysis: two-tailed student’s t-test (pTT). Statistic source data are found in Supplementary Table 3.Unprocessed blots are shown in Supplementary Fig. 11.
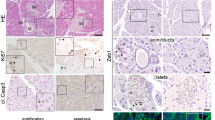
Supplementary Figure 6 Inhibition of EGFR, PI3K and aPKC signaling during differentiation of hESCs into β-cells affects NGN3 expression and β-cell number.
(a) Quantification of the fraction of NGN3+ cells (Number of NGN3 cells/DAPI area) in cultures of Ctrl, LY-, aPKCi-, or EGFRi-treated (24 h) hESC-derived NGN3+ cells (EPd4-Protocol A). n = 3 wells of cells, from three independent differentiations, Ctrl/LY pTT = 0.2892, Ctrl/aPKCi pTT = 0.1020, Ctrl/EGFRi pTT = 0.5611. (b) Quantification of the fraction of NGN3+ cells (Number of NGN3 cells/DAPI area) in cultures of Ctrl, LY-, aPKCi-, or EGFRi- or LY + aPKCi-treated (24 h) hESC-derived NGN3+ cells (S4d9-Protocol B). n = 3 wells of cells, from three independent differentiations, Ctrl/LY pTT = 0.5421, Ctrl/aPKCi pTT = 0.6711, Ctrl/EGFRi pTT = 0.8018, Ctrl/LY + aPKCi pTT = 0.6893. (c) Representative images of cultures of hESC-derived NGN3+ cells (S4d9-Protocol B), treated for 24 h with or without LY, aPKCi, EGFRi or LY + aPKCi and stained for NGN3 (red) and DAPI (blue). (d) Representative images of cultures of hESC-derived β-cells (S6d16-Protocol B), treated for 1 week with or without LY, aPKCi, EGFRi or LY + aPKCi and stained for C-peptide (white) and DAPI (blue). (e) Mean NGN3 intensity quantification in NGN3+ cells in cultures of Ctrl, LY-, aPKCi-, or EGFRi- or LY + aPKCi-treated (24 h) hESC-derived NGN3+ cells (S4d9-Protocol B). n = 4 wells of cells for each condition, (Total number of cells: 1723, 2103, 1946, 1987, 2065 (from left to right)), from 4 independent differentiations, Ctrl/LY pKW = 0.043, Ctrl/aPKCi pKW = 0.021, Ctrl/EGFRi pKW = 0.021, Ctrl/LY + aPKCi pKW = 0.021. (f) β-cell differentiation (relative C-peptide (C-pep/DAPI) compared to Ctrl) quantification in cultures of Ctrl, LY-, aPKCi-, or EGFRi- or LY + aPKCi-treated (7 days) hESC-derived β-cells (S6d16-Protocol B). n = 4 wells of cells, from 4 independent differentiations, Ctrl/LY pTT = 0.0023, Ctrl/aPKCi pTT = 0.0009, Ctrl/EGFRi pTT = 0.0245, Ctrl/LY + aPKCi pTT = 0.0163, LY/LY + aPKCi pTT = 0.0002 (g) Representative images of cultures of hESC-derived β-cells (EPd10-Protocol A), treated for 7 days with or without LY, aPKCi or EGFRi were stained for C-peptide (C-pep; white) NKX6-1 (red), PDX1 (green) and DAPI (blue). Error bars represent ± SEM. Scale bars, 50 μm. Statistic source data can be found in Supplementary Table 3. Statistical analysis: two-tailed student’s t-test (pTT) or Kruskal-Wallis (pKW).
Supplementary Figure 7 Generation of a human embryonic stem cell based NGN3GFP reporter line.
(a) The strategy for targeting the NGN3 locus. The NGN3 targeting vector was inserted into the 5’-untranslated region of the NGN3 gene, upstream of the NGN3 start codon (ATG), resulting in an GFP-tagged NGN3 allele. (b) Representative 3D-reconstruction z-stack of hESC-derived EPd3 cells differentiated using protocol A, stained for GFP, EZRIN and DAPI. Right panel represents the culture viewed from the side. (c) Representative image of hESC-derived EPd4 cells differentiated using protocol A, stained for NGN3, GFP and DAPI to show that the reporter is functional. (d) Representative flow cytometry plot of GFP expression of the NGN3GFP reporter differentiated to EPd2 and EPd9 using protocol A. (e) Percentage (%) of GFP expressing cells in Control (untagged SA121 cells) versus NGN3GFP cell lines at EPd2-EPd9 differentiated using protocol A. n = 2 except d9 n = 1. (f) qPCR analysis of GFP from PEd11-EPd10 cultures of human embryonic stem cells differentiated to endocrine cells using protocol A, or S3d7-S5d13 cultures differentiated using protocol B. n = 1. (g) qPCR analysis of NGN3 from PEd11-EPd10 cultures of human embryonic stem cells differentiated to endocrine cells using protocol A, or S3d7-S5d13 cultures differentiated using protocol B. n = 1. (h) qPCR analysis of INS from PEd11-EPd10 cultures of human embryonic stem cells differentiated to endocrine cells using protocol A, or S3d7-S5d13 cultures differentiated using protocol B. n = 1. (i) Ngn3High/NGN3High and Ngn3Low/NGN3Low cells were isolated using flow cytometry from E15.5 Ngn3Y FP+ pancreata or hESC-derived NGN3+ cells (EPd3—Protocol A). Expression of Ngn3, Muc1, Sox9, Hes1, Pdx1, Isl-1 and NeuroD was determined by qPCR and normalized to β-actin. Mouse: n = 5 extracts of RNA (from 1 pancreata each), from three independent experiments, Ngn3 pTT = 0.0414, Muc1 pTT = 0.0066, Sox9 pTT = 0.0289, Hes1 pTT = 0.0286, Pdx1 pTT = 0.7503, Isl-1 pTT = 0.0106, NeuroD pTT = 0.0411; Human: n = 3 extracts of RNA (from 1 differentiated plate each), from three independent differentiations, NGN3 pTT = 0.0127, MUC1 pTT = 0.0461, SOX9 pTT = 0.0102, HES1 pTT = 0.0117, PDX1 pTT = 0.2470, ISL1 pTT = 0.0184, NEUROD pTT = 0.0301. Error bars represent ± SEM. Scale bar, 100 μm (b), 20 μm (c). Statistic source data can be found in Supplementary Table 3. Statistical analysis: two-tailed student’s t-test (pTT).
Supplementary Figure 8 Inhibition of EGFR, PI3K or aPKC signaling during differentiation of hESCs into β-cells affects apical polarity.
(a–c) qPCR analysis of Prkciota (a), Prkczeta (b) or NGN3 (c) in lysates from cultures of hESC-derived NGN3+ cells (EPd4—protocol A), after 24 h siRNA knockdown (KD) of aPKCiota (Prkci KD), aPKCzeta (Prkcz KD) or aPKCiota + aPKCzeta simultaneously (Prkci+z KD), compared to Ctrl. n = 6, 6, 6, 3 wells of cells (from left to right), from two independent experiments, (a) Ctrl/Prkci pTT < 0.0001, Ctrl/Prkcz pTT = 0.4520 (n.s.), Ctrl/Prkci + z pTT = 0.0055. (b) Ctrl/Prkci pTT = 0.7266 (n.s.), Ctrl/Prkcz pTT < 0.0001, Ctrl/Prkci + z pTT < 0.0001. (c) Ctrl/Prkci pTT = 0.0377, Ctrl/Prkcz pTT = 0.0453, Ctrl/Prkci + z pTT = 0.00177. (d) Quantification of the relative number of NGN3 + cells in cultures of EPd4 cells (protocol A), after 24 h siRNA knockdown (KD) of aPKCiota (Prkci KD), aPKCzeta (Prkcz KD) or aPKCiota + aPKCzeta simultaneously (Prkci+z KD) were compared to Ctrl. n = 3 wells of cells, from two independent differentiations, Ctrl/Prkci pTT = 0.6861, Ctrl/Prkcz pTT = 0.5007, Ctrl/Prkci + z pTT = 0.4540. (e) Representative cultures of hESC-derived NGN3 + cells (EPd3—Protocol A), stained for E-cadherin (E-cad;red), GFP (green) and DAPI (blue). Inset shows magnification of indicated region. (f) Representative cultures of hESC-derived NGN3 + cells (EPd3—Protocol A), stained for F-actin (red), GFP (green) and DAPI (blue). Inset shows magnification of indicated region. (g) Representative 3D-reconstruction of z-stack of a culture of hESC-derived NGN3 + cells (EPd3—Protocol A), stained for ZO-1 (red), GFP (green), Ezrin (white) and DAPI (blue). Inset shows magnification of indicated region. Right panel represents the culture viewed from the side. (h–j) qPCR analysis of aPKCiota (h), Muc1 (i) and Hes1 (j) in flow cytometry isolated Ctrl, LY-, aPKCi- or EGFRi-treated (24 h) hESC-derived NGN3 + cells (EPd4—protocol A). n = 3 extracts of RNA (from 1 differentiated plate each), from three independent differentiations, (h) Ctrl/LY pTT = 0.0444, Ctrl/aPKCi pTT = 0.0238, Ctrl/EGFRi pTT = 0.0437 (i) Ctrl/LY pTT = 0.0196, Ctrl/aPKCi pTT = 0.0013, Ctrl/EGFRi pTT = 0.0257 (j) Ctrl/LY pTT = 0.0178, Ctrl/aPKCi pTT = 0.0392, Ctrl/EGFRi pTT = 0.424. (k–m) Whole lysates from EPd10 cultures (protocol A), treated with LY, aPKCi or EGFRi for 7 days, were analyzed for the expression of Ezrin and aPKC using western blot. alpha-tubulin was used as loading control. Relative Band intensity show that LY and EGFRi treatment increase the expression of Ezrin and aPKC, whereas aPKCi treatment decreases them. n = 2. Error bars represent ± SEM. Scale bars, 50 μm (e–g). Statistic source data are found in Supplementary Table 3. Statistical analysis: two-tailed student’s t-test (pTT).
Supplementary information
Supplementary Information
Supplementary Information (PDF 20649 kb)
Supplementary Table 1
Supplementary Information (XLSX 33 kb)
Supplementary Table 2
Supplementary Information (XLSX 11 kb)
Supplementary Table 3
Supplementary Information (XLSX 337 kb)
Dynamics in Ngn3 expression and apical membrane length.
Time-lapse recording of Ngn3RG1/Muc1mCherry pancreatic explants of the conversion of Ngn3Low to Ngn3High cells revealed that Ngn3Low cells (marked with a dot) are fully polarized and located in close proximity to the primitive duct lumen. Concomitant with apical domain size reduction and increased Ngn3 expression (Ngn3High), the cell body moves to a basal position within the epithelium. (AVI 653 kb)
Conversion from Ngn3Low to Ngn3High in WT.
Time-lapse recording of Ngn3RG1/Muc1mCherry pancreatic explants in the WT revealed that the time to convert Ngn3Low cells (RFP +) into Ngn3High cells (RFP + /GFP +) was ≈ 4 h (from birth until the cell (marked with a dot) turns on GFP). Quantifications can be found in Supplementary Fig. 3p. Statistic source data can be found in Supplementary Table 3. (AVI 1203 kb)
Conversion from Ngn3Low to Ngn3High after LY treatment.
Time-lapse recording of Ngn3RG1/Muc1mCherry pancreatic explants in the presence of LY revealed that the time to convert Ngn3Low cells (RFP +) into Ngn3High cells (RFP + /GFP +) was ≈ 8 h (from birth until the cell (marked with a dot) turns on GFP). Quantifications can be found in Supplementary Fig. 3p. Statistic source data can be found in Supplementary Table 3. (AVI 1208 kb)
EZRIN expression in the culture of hESC-derived NGN3 cells.
Immunofluorescence stainings with an antibody against the apical marker EZRIN revealed that the epithelial cells facing the media were polarized. EZRIN (white), DAPI (blue) & GFP (green). (AVI 1591 kb)
Rights and permissions
About this article
Cite this article
Löf-Öhlin, Z., Nyeng, P., Bechard, M. et al. EGFR signalling controls cellular fate and pancreatic organogenesis by regulating apicobasal polarity. Nat Cell Biol 19, 1313–1325 (2017). https://doi.org/10.1038/ncb3628
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3628
This article is cited by
-
Synaptotagmin-13 orchestrates pancreatic endocrine cell egression and islet morphogenesis
Nature Communications (2022)
-
Engineering islets from stem cells for advanced therapies of diabetes
Nature Reviews Drug Discovery (2021)
-
HDAC6 promotes growth, migration/invasion, and self-renewal of rhabdomyosarcoma
Oncogene (2021)
-
A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion
Nature Communications (2021)
-
Cell polarity and oncogenesis: common mutations contribute to altered cellular polarity and promote malignancy
The Nucleus (2020)