Abstract
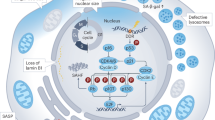
Senescence, a persistent form of cell-cycle arrest, is often associated with a diverse secretome, which provides complex functionality for senescent cells within the tissue microenvironment. We show that oncogene-induced senescence is accompanied by a dynamic fluctuation of NOTCH1 activity, which drives a TGF-β-rich secretome, while suppressing the senescence-associated pro-inflammatory secretome through inhibition of C/EBPβ. NOTCH1 and NOTCH1-driven TGF-β contribute to ‘lateral induction of senescence’ through a juxtacrine NOTCH–JAG1 pathway. In addition, NOTCH1 inhibition during senescence facilitates upregulation of pro-inflammatory cytokines, promoting lymphocyte recruitment and senescence surveillance in vivo. As enforced activation of NOTCH1 signalling confers a near mutually exclusive secretory profile compared with typical senescence, our data collectively indicate that the dynamic alteration of NOTCH1 activity during senescence dictates a functional balance between these two distinct secretomes: one representing TGF-β and the other pro-inflammatory cytokines, highlighting that NOTCH1 is a temporospatial controller of secretome composition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Kuilman, T. & Peeper, D. S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9, 81–94 (2009).
Childs, B. G., Durik, M., Baker, D. J. & van Deursen, J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015).
Acosta, J. C. et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 15, 978–990 (2013).
Hubackova, S., Krejcikova, K., Bartek, J. & Hodny, Z. IL1- and TGFβ-Nox4 signaling, oxidative stress and DNA damage response are shared features of replicative, oncogene-induced, and drug-induced paracrine ‘Bystander senescence’. Aging (Albany NY) 4, 932–951 (2012).
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Kang, T.-W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
Krtolica, A., Parrinello, S., Lockett, S., Desprez, P. Y. & Campisi, J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl Acad. Sci. USA 98, 12072–12077 (2001).
Coppé, J.-P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Kopan, R. & Ilagan, M. X. G. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009).
Ntziachristos, P., Lim, J. S., Sage, J. & Aifantis, I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25, 318–334 (2014).
Weekes, M. P. et al. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340, 199–202 (2013).
Young, A. R. J. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009).
Cui, H., Kong, Y., Xu, M. & Zhang, H. Notch3 functions as a tumor suppressor by controlling cellular senescence. Cancer Res. 73, 3451–3459 (2013).
Kagawa, S. et al. Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities. Oncogene 34, 2347–2359 (2015).
Procopio, M.-G. et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat. Cell Biol. 17, 1193–1204 (2015).
Kirschner, K. et al. Phenotype specific analyses reveal distinct regulatory mechanism for chronically activated p53. PLoS Genet. 11, e1005053 (2015).
Acosta, J. C. et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006–1018 (2008).
Orjalo, A. V., Bhaumik, D., Gengler, B. K., Scott, G. K. & Campisi, J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc. Natl Acad. Sci. USA 106, 17031–17036 (2009).
Fryer, C. J., Lamar, E., Turbachova, I., Kintner, C. & Jones, K. A. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 16, 1397–1411 (2002).
Lee, B. Y. et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell 5, 187–195 (2006).
Hata, A., Lo, R. S., Wotton, D., Lagna, G. & Massague, J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature 388, 82–87 (1997).
Lewis, J. Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell Dev. Biol. 9, 583–589 (1998).
Hartman, B. H., Reh, T. A. & Bermingham-McDonogh, O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc. Natl Acad. Sci. USA 107, 15792–15797 (2010).
D’Souza, B., Miyamoto, A. & Weinmaster, G. The many facets of Notch ligands. Oncogene 27, 5148–5167 (2008).
Kurpinski, K. et al. Transforming growth factor-beta and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells 28, 734–742 (2010).
Collado, M. et al. Tumour biology: senescence in premalignant tumours. Nature 436, 642 (2005).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Miyamoto, Y. et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 3, 565–576 (2003).
Avila, J. L. & Kissil, J. L. Notch signaling in pancreatic cancer: oncogene or tumor suppressor? Trends Mol. Med. 19, 320–327 (2013).
Caldwell, M. E. et al. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer. Oncogene 31, 1599–1608 (2011).
Lee, W.-Y. & Kubes, P. Leukocyte adhesion in the liver: distinct adhesion paradigm from other organs. J. Hepatol. 48, 504–512 (2008).
Shetty, S., Weston, C. J., Adams, D. H. & Lalor, P. F. A flow adhesion assay to study leucocyte recruitment to human hepatic sinusoidal endothelium under conditions of shear stress. http://dx.doi.org/10.3791/51330 J. Vis. Exp. (2014).
Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011).
Huggins, C. J. et al. C/EBPγ suppresses senescence and inflammatory gene expression by heterodimerizing with C/EBPβ. Mol. Cell. Biol. 33, 3242–3258 (2013).
Nakazato, Y. et al. Interleukin (IL)-1 and IL-4 synergistically stimulate NF-IL6 activity and IL-6 production in human mesangial cells. Kidney Int. 54, 71–79 (1998).
ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Storer, M. et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119–1130 (2013).
Muñoz-Espín, D. et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013).
Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015).
Boni, A. et al. Notch1 regulates the fate of cardiac progenitor cells. Proc. Natl Acad. Sci. USA 105, 15529–15534 (2008).
Pitt, L. A. et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell 27, 755–768 (2015).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Dörr, J. R. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013).
Yagüe, E. et al. Ability to acquire drug resistance arises early during the tumorigenesis process. Cancer Res. 67, 1130–1137 (2007).
Narita, M. et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 (2006).
Aziz, N., Cherwinski, H. & McMahon, M. Complementation of defective colony-stimulating factor 1 receptor signaling and mitogenesis by Raf and v-Src. Mol. Cell. Biol. 19, 1101–1115 (1999).
Capobianco, A. J., Zagouras, P., Blaumueller, C. M., Artavanis-Tsakonas, S. & Bishop, J. M. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 17, 6265–6273 (1997).
Cox, J. et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 (2009).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Gonzalez, R. et al. Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc. Natl Acad. Sci. USA 107, 3552–3557 (2010).
Sadaie, M. et al. Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 27, 1800–1808 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Shetty, S. et al. Common lymphatic endothelial and vascular endothelial receptor-1 mediates the transmigration of regulatory T cells across human hepatic sinusoidal endothelium. J. Immunol. 186, 4147–4155 (2011).
Lalor, P. F. et al. Vascular adhesion protein-1 mediates adhesion and transmigration of lymphocytes on human hepatic endothelial cells. J. Immunol. 169, 983–992 (2002).
Pruitt, K. D. et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763 (2014).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Schmidt, D. et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 (2009).
Sargent, J. L. et al. A TGFβ-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J. Invest. Dermatol. 130, 694–705 (2010).
Brennan, E. P. et al. Next-generation sequencing identifies TGF-β1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim. Biophys. Acta 1822, 589–599 (2012).
Acknowledgements
We are very grateful to J. Gil (Imperial College London, UK), A. Harris (CRUK, Oxford, UK) and D. Peeper (NKI, Amsterdam) for reagents; J. Miller and L.-A. McDuffus for assistance with the immunohistochemistry; E. Serrao and K. Brindle (CRUK Cambridge Institute, UK) for the murine pancreatic tissue; G. Slater for analysis of the IL1A locus; M. Clayton for the tail-vein injections; and A. Young and M. Narita for technical support. This work was supported by the University of Cambridge, Cancer Research UK and Hutchison Whampoa. The M.N. laboratory is supported by Cancer Research UK Cambridge Institute Core Grant (C14303/A17197). M.H. was supported by CRUK Translational Medicine Research Fellowship and CRUK Clinician Scientist Fellowship (C52489/A19924). This work was also supported by a Wellcome Trust PRF (WT101835) to P.J.L., a Wellcome Trust Senior Fellowship to M.P.W. (108070/Z/15/Z), a Wellcome Trust Training Fellowship to N.J.M. (093964/Z/10/Z), and a Wellcome Trust Intermediate Fellowship (097162/Z/11/Z) to S.S. L.Z. was funded by the German Research Foundation (DFG; grants FOR2314 and SFB685), the Gottfried Wilhelm Leibniz Program, the European Research Council (projects ‘CholangioConcept’), the German Ministry for Education and Research (BMBF) (eMed-Multiscale HCC), the German Universities Excellence Initiative (third funding line: ‘future concept’), the German Center for Translational Cancer Research (DKTK) and the German–Israeli Cooperation in Cancer Research (DKFZ–MOST).
Author information
Authors and Affiliations
Contributions
M.H. and M.N. designed the experiments; M.H., Y.I. and A.J.P. carried out the in vitro experiments; M.P.W., N.J.M. and R.A. performed and analysed the proteomics experiments; M.H. and T.-W.K. performed and analysed the in vivo experiments; D.A.P. and S.S. performed the HSEC experiments; S.M. and R.S. performed bioinformatic analysis; K.T. provided reagents; W.H. performed the analysis of immunohistochemistry images. P.J.L., L.Z. and M.N. provided resources; M.H. and M.N. wrote the manuscript; all authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Plasma membrane proteomics (PMP) defines the cell surface signature of senescence including NOTCH1.
(a) ER:HRASG12V IMR90 cells with RAS-induced senescence (RIS), induced by 6 days of 4OHT or DNA-damage induced senescence (DDIS) induced by Etoposide, were analysed for SA-β-GAL staining (upper panels and lower left), presence of senescence-associated heterochromatic foci (SAHF) and DNA synthesis by BrdU incorporation (lower right). Manual counting of >200 cells; bars are means; unpaired t-test; n = 3 biologically independent experiments; ∗∗∗P ≤ 0.001. Scale bar 200 μm. (b) Correlation of log2 fold expression changes of 521 high-confidence PMP ‘hits’ at cell surface protein level by PMP (x axis) with transcriptomic changes by mRNA-Seq (y axis). Hits are colour-coded by significance level: green dots indicate a proteomic significance of P < 0.05; yellow indicate a transcript q < 0.01; red indicates significant in both PMP and transcript datasets. Correlation was calculated for all 521 (all), those with significance at transcript or protein level (anySig) or both transcript and protein (Commsig). FDR, false discovery rate. (c) Network enrichment analysis, with networks defined by the presence of a PMP hit and populated by transcript level data revealed the highest enriched network contained NOTCH1 as a major network hub. Other network members are shown by subcellular localisation; red indicates upregulation and green downregulation. Among four NOTCH receptors, only NOTCH1 was significantly upregulated in both PM proteomic and transcriptomic data from RIS cells. (d) ESF and WI38 human diploid fibroblast cell lines stably expressing oncogenic HRASG12V were analysed for cell surface NOTCH1 expression by flow cytometry. (e) ΔMEK1:ER IMR90 cells with and without induction by 4OHT were analysed for cell surface NOTCH1 expression by flow cytometry. Statistics source data for A are provided in Supplementary Table 2.
Supplementary Figure 2 Dynamic canonical NOTCH1 signalling is responsible for reciprocal regulation of TGF-β ligands and proinflammatory cytokines during senescence.
(a) ER:HRASG12V IMR90 cells were examined in a time series analysis of TGF-β ligand expression by qRT-PCR (upper) and JAG1 expression by immunoblotting (lower) after RAS induction by 4OHT; values are mean ± s.e.m.; the experiment was performed four times for day 0 through 6 and three times for day 8 with similar results. (b) ER:HRASG12V IMR90 cells with and without 3 days treatment with 4OHT and DAPT at the indicated concentrations were analysed by qRT-PCR; One-way ANOVA with Dunnett’s multiple comparison test; values are mean ± s.e.m.; n = 6 biologically independent experiments for HES1, IL1A and IL8, n = 5 biologically independent experiments for TGFB1. ∗P ≤ 0.05, ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001. (c) Time-dependent behaviour of TRE-N1ICD-FLAG IMR90 cells after induction of N1ICD expression by addition of doxycycline: immunoblot demonstrating expression of indicated proteins. (d) TRE-N1ICD IMR90 cells with and without canonical NOTCH-inhibitor dnMAML1 were analysed for colony forming capacity. (e) TRE-N1ICD IMR90 cells treated with or without doxycycline for 7 days were then either cultured with continued doxycycline treatment or in regular media for a further 10 days before analysis of colony forming capacity. Statistics source data for a and b are provided in Supplementary Table 2.
Supplementary Figure 3 Activation of NOTCH is associated with cell-cycle arrest and antagonism of the RAS-driven secretome.
(a) Multiple cell-cycle-related genesets were significantly enriched within downregulated genes from IMR90 cells with N1ICD- (NIS), HRASG12V-(RIS), or Etoposide-induced senescence (DDIS). Upper: example Geneset enrichment analysis (GSEA) plots for the ‘Reactome–cell cycle’ geneset in the indicated senescence dataset (NES, normalised enrichment score; FDR, false discovery rate); lower: table demonstrating enrichment of multiple cell-cycle genesets in down-regulated genes. (b) GSEA demonstrating enrichment of publically available TGF-β1-signatures in transcriptomic data from IMR90 cells stably expressing N1ICD (NIS). (c) Differentially expressed transcripts in cells expressing N1ICD-, HRASG12V-or both (NIS, RIS, or N+RIS, respectively), compared to normal control cells. Heat map shows z-score normalised fold changes of 1150 secretome genes differentially expressed in at least in one comparison. Representative KEGG pathways enriched in five clusters (FDR < 0.01) are shown. Note, GLB1 encodes the lysosomal enzyme responsible for SA-β-Gal activity.
Supplementary Figure 4 NOTCH1 drives autonomous and non-autonomous cell-cycle arrest in part through TGF-β.
(a) Left: proliferation analysis of TRE-N1ICD IMR90 cells treated with or without doxycycline with or without the TGF-β receptor antagonist GW788388. Right: The proliferative ability of mRFP IMR90 cells was analysed during co-culture with TRE-N1ICD IMR90 cells treated with or without doxycycline and GW788388; Values are mean ± s.e.m., representative of three independent experiments with similar results. (b) Left: proliferation analysis of TRE-N1ICD IMR90 cells with dnSMAD4-mVenus or vector control treated with or without doxycycline. Middle: The proliferative ability of mRFP IMR90 cells was analysed during co-culture with TRE-N1ICD IMR90 cells with dnSMAD4-mVenus or vector control treated with or without doxycycline; Values are mean ± s.e.m., representative of three independent experiments with similar results. Right: Immunofluorescence for TGFBI in IMR90 cells with dnSMAD4-mVenus or vector control treated with 10 ng ml−1 TGF-β1 for 8 h. Scale bar 30 μm. (c) IMR90 cells were subject to short-term proliferation analysis with or without the indicated concentrations of recombinant TGF-β1, 2 or 3; values are mean ± s.e.m.; representative of three independent experiments with similar results. (d) Representative immunofluorescence of SMAD2/3 after 24 h treatment with 10 ng ml−1 TGF-β1, 2, 3 or vehicle. Scale bar 50 μm.
Supplementary Figure 5 NOTCH1 drives juxtacrine senescence through JAG1-mediated lateral induction.
(a) IMR90 cells stably expressing N1ICD underwent transcriptional profiling by mRNA-seq; differential expression of canonical NOTCH ligands was then analysed. (b) Immunoblot for JAG1 and coomassie gel staining of whole cell lysate (WCL) and conditioned media (CM) from TRE-N1ICD IMR90 cells treated with or without doxycycline for 3 days. (c) RPE1 cells expressing ectopic N1ICD were analysed for markers of senescence; expression of the indicated proteins by immunoblotting; SA-β-gal and DNA synthesis by BrdU incorporation; unpaired t-test; values are mean ± s.e.m.; n = 6 biologically independent experiments; ∗∗∗P ≤ 0.001 relative to vector control. Insets, magnified picture of dotted rectangular areas. Scale bar 200 μm. (d) Analysis of proliferation, by BrdU incorporation, and expression of p21 by immunofluorescence in mRFP-IMR90 cells when cocultured with TRE-N1ICD IMR90 cells with vector or shJAG1-4 and with or without 10 μM DAPT (See analogous experiment in Fig. 5bg). Bars are means of ≥200 mRFP-positive cells per condition; n = 3 biologically independent experiments; one-way ANOVA with Dunnett’s multiple comparisons test; ∗∗P ≤ 0.01, ∗∗∗P ≤ 0.001. (e) The proliferative ability of RPE1 cells expressing N1ICD or vector was analysed (upper). The proliferative ability of mRFP-IMR90 cells during co-culture with N1ICD or vector-expressing RPE1 cells, with and without DAPT, was analysed (lower) by proliferation assay; values are mean ± s.e.m.; representative of three independent experiments with similar results. (f) Model of NOTCH-induced lateral induction of NOTCH-signalling through JAG1 and TGF-β. How exactly the NOTCH activation induces JAG1 and TGF-β remains to be elucidated (dotted line). Activation of NOTCH signalling in the left-hand cell leads to the expression of both JAG1 and TGF-β. JAG1 acts in a cell-contact dependent fashion to induce activation of endogenous NOTCH in the right-hand cell and in concert with TGF-β to induce juxtacrine senescence. (g) Proliferation of mRFP1-IMR90 cells was analysed during co-culture with TRE-N1ICD cells treated with or without doxycycline and ER:RAS cells expressing dnMAML1-mVenus treated with or without 4OHT; representative of three independent experiments with similar results. Statistics source data for a,c and d are provided in Supplementary Table 2.
Supplementary Figure 6 Notch1 is upregulated during in vivo Ras-induced senescence (RIS) and mediates lateral induction of senescence and Notch signalling.
(a) Pancreatic tissue from p48-cre; LSL-KrasG12D and matched control mice was analysed by immunohistochemistry (IHC) for Dec1 and Notch1; insets demonstrate the staining pattern within normal pancreatic ducts (upper, arrowhead), pancreatic intraepithelial neoplasia (PanIN) (middle) and acinar-ductal metaplasia (ADM) (lower). Scale bar 200 μm. (b) Time series analysis of serial sections of murine liver tissue 9 days after HDTV-injection of NRASG12V-IRES-mVenus or NRASG12V-IRES-dnMAML1-mVenus stained by IHC for NRAS and Hes1; quantitation of positivity for Hes1 in NRAS-positive cells; bars are means; unpaired t-test; n = 3 mice per time point; ∗P ≤ 0.05. (c) Representative images of two-colour IHC staining of murine liver sections 9 days after HDTV-injection of NRASG12V-IRES-mVenus for NRAS (brown) and Hes1 (red) demonstrating Hes1-positive/ NRAS-negative cells (asterisk) adjacent to NRAS-positive hepatocytes. Arrows demonstrate Hes1-negative cells for comparison. Scale bar 50 μm. (d,e) IHC for NRAS and p21 on serial murine liver sections at indicated time points after HDTV injection of NRASG12V-IRES-mVenus or NRASG12V-IRES-dnMAML1-mVenus. Representative IHC images; Insets, magnified picture of dotted rectangular areas. Asterisk demonstrates p21-positive/NRAS-negative cells adjacent to NRAS-positive hepatocytes (d). Scale bar 200 μm. Quantitation of p21 expression in NRAS-positive and negative hepatocyte populations (e). Hepatocytes were dichotomised into NRAS-positive (top) or negative (bottom) populations by automated image analysis and then p21 expression analysed in these 2 populations; values are mean ± s.e.m. from automated image analysis of ≥105 cells per liver section; for NRASG12V injected animals at D6, 9 & 12, n = 3, 3 & 4 mice respectively; for NRASG12V-IRES-dnMAML1 injected animals at D6, 9 & 12, n = 4, 3 & 5 mice respectively; unpaired t-test. (f) Representative immunohistochemistry (upper) and immunofluorescence (lower) of CD3-expressing T-lymphocytes within the livers of mice injected with either NRASG12V-IRES-mVenus or NRASG12V-IRES-dnMAML1-mVenus. Arrows indicate CD3+ T-lymphocytes. (See also Fig. 6c) Scale bar upper panels 200 μm, lower panels 25 μm. (g) Quantification of B220 (B-lymphocyte marker) positive cells within indicated livers as in Fig. 6c. Values are mean ± s.e.m. from automated image analysis of ≥105 cells from liver section; for NRASG12Vinjected animals at D6, 9 & 12, n = 3, 3 & 4 mice respectively; for NRASG12V-IRES-dnMAML1 injected animals at D6, 9 & 12, n = 4, 3 & 5 mice respectively. Statistics source data for b,e and g are provided in Supplementary Table 2.
Supplementary Figure 7 Experimental setup for human sinusoidal endothelial cell (HSEC) flow-adhesion assay.
(a) Workflow for production of conditioned media (CM) from ER:HRASG12V/TRE-N1ICD IMR90 cells with induction timing of HRAS and N1ICD and final incubation in serum free media for 16 h before harvesting; similar schema for ER:HRASG12V IMR90s with or without dnMAML1 and harvesting at d3 of HRAS expression. (b) Diagram illustrating the set-up of the flow adhesion assay: HSEC from human liver tissue were plated within the flow cell and cultured until confluent, before incubation for 24 h in differentially conditioned media. Then peripheral blood lymphocytes (PBLs) from healthy individuals were perfused over the HSEC in fresh media and analysed for their ability to adhere to the HSEC.
Supplementary Figure 8 NOTCH1 controls the pro-inflammatory SASP through repression of C/EBPβ.
(a) IMR90 cells expressing HRASG12V, N1ICD-FLAG or both were analysed for expression of CEBPB by qRT-PCR; One-way ANOVA with Dunnett’s multiple comparison test; values are mean ± s.e.m.; n = 6 biologically independent experiments; ∗∗∗P ≤ 0.001. (b) Genes upregulated by C/EBPβ are highly enriched by geneset enrichment analysis amongst genes upregulated by HRASG12V (RIS) and downregulated by N1ICD (NIS) and the co-expression of N1ICD and HRASG12V (N+RIS); FDR, false discovery rate; NES, normalised enrichment score. (c) TRE-N1ICD IMR90 cells treated with or without doxycycline for 3 days, then with or without 100 ng ml−1 TNF-α for 1 h were analysed for expression and chromatin binding of indicated NF-κB family components by immunoblot. (d) Ectopic expression of N1ICD-FLAG in HaCaT immortalised human keratinocyte cells leads to a downregulation of C/EBPβ, but not cell-autonomous proliferative arrest; analysis by immunoblot and qRT-PCR; bars are means; unpaired t-test; n = 3 biologically independent experiments; ∗∗∗P ≤ 0.001. (e) Genome browser snapshot of the IL1A locus demonstrating ChIP-Seq binding profiles for C/EBPβ, key histone marks and accessibility of chromatin by DNAse-Seq (derived from ENCODE). There is a small C/EBPβ binding peak at the Gencode-annotated TSS (proximal), but much larger binding peaks at a distal enhancer region 8kb upstream (distal), recently demonstrated to be tethered to the IL1A core promoter by 3D chromatin interactome analysis. Representative RefSeq and Gencode annotated IL1A transcripts are shown. Solid vertical lines represent C/EBPβ ChIP-qPCR ‘target’ sites at the ‘Proximal’ and ‘Distal’ regions, whereas the dotted vertical line represents ‘non-specific’ site. (f) Chromatin immunoprecipitation of endogenous C/EBPβ (from experiment described in Fig. 8g) and subsequent qPCR for loci at IL6 and IL8 promoters; n = 3 biologically independent experiments; one way ANOVA with Dunnett’s multiple comparison test; values are mean ± s.e.m. ∗∗∗P ≤ 0.001. Statistics source data for a,d and f are provided in Supplementary Table 2.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2144 kb)
Supplementary Table 1
Supplementary Information (XLSX 100 kb)
Supplementary Table 2
Supplementary Information (XLSX 218 kb)
Rights and permissions
About this article
Cite this article
Hoare, M., Ito, Y., Kang, TW. et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat Cell Biol 18, 979–992 (2016). https://doi.org/10.1038/ncb3397
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3397
This article is cited by
-
Modulation of fracture healing by senescence-associated secretory phenotype (SASP): a narrative review of the current literature
European Journal of Medical Research (2024)
-
Therapeutic targeting of cellular senescence in diabetic macular edema: preclinical and phase 1 trial results
Nature Medicine (2024)
-
RSK3 switches cell fate: from stress-induced senescence to malignant progression
Journal of Experimental & Clinical Cancer Research (2023)
-
Endothelial senescence in vascular diseases: current understanding and future opportunities in senotherapeutics
Experimental & Molecular Medicine (2023)
-
SLC35D3 promotes white adipose tissue browning to ameliorate obesity by NOTCH signaling
Nature Communications (2023)