Abstract
Multiple lines of evidence indicate that mitochondrial dysfunction is central to Parkinson’s disease. Here we investigate the mechanism by which parkin, an E3 ubiquitin ligase, and USP30, a mitochondrion-localized deubiquitylase, regulate mitophagy. We find that mitochondrial damage stimulates parkin to assemble Lys 6, Lys 11 and Lys 63 chains on mitochondria, and that USP30 is a ubiquitin-specific deubiquitylase with a strong preference for cleaving Lys 6- and Lys 11-linked multimers. Using mass spectrometry, we show that recombinant USP30 preferentially removes these linkage types from intact ubiquitylated mitochondria and counteracts parkin-mediated ubiquitin chain formation in cells. These results, combined with a series of chimaera and localization studies, afford insights into the mechanism by which a balance of ubiquitylation and deubiquitylation regulates mitochondrial homeostasis, and suggest a general mechanism for organelle autophagy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Nunnari, J. & Suomalainen, A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012).
Chan, D. C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 46, 265–287 (2012).
Ashrafi, G. & Schwarz, T. L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42 (2013).
Saiki, S., Sato, S. & Hattori, N. Molecular pathogenesis of Parkinson’s disease: update. J. Neurol. Neurosurg. Psychiatry 83, 430–436 (2012).
Kubli, D. A. & Gustafsson, Å. B. Mitochondria and mitophagy: the yin and yang of cell death control. Circ. Res. 111, 1208–1221 (2012).
Vincow, E. S. et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl Acad. Sci. USA 110, 6400–6405 (2013).
Narendra, D., Walker, J. E. & Youle, R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb. Perspect. Biol. 4, 23125018 (2012).
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 (2010).
Narendra, D., Tanaka, A., Suen, D-F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Jin, S. M. et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 (2010).
Chan, N. C. et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726–1737 (2011).
Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013).
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and Ubiquitin chain synthesis. Mol. Cell 25284222 (2014).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Valente, E. M. et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 56, 336–341 (2004).
Komander, D. & Clague, M. J. Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Bingol, B. et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375 (2014).
Wang, X. et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 (2011).
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 (2010).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Phu, L. et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell Proteomics 10, M110.003756 (2011).
Bozza, W. P., Liang, Q., Gong, P. & Zhuang, Z. Transient kinetic analysis of USP2-catalyzed deubiquitination reveals a conformational rearrangement in the K48-linked diubiquitin substrate. Biochemistry 51, 10075–10086 (2012).
Zhang, W. et al. Contribution of active site residues to substrate hydrolysis by USP2: insights into catalysis by ubiquitin specific proteases. Biochemistry 50, 4775–4785 (2011).
Rogov, V., Dötsch, V., Johansen, T. & Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178 (2014).
Catic, A. et al. Screen for ISG15-crossreactive deubiquitinases. PLoS ONE 2, e679 (2007).
Malakhov, M. P., Malakhova, O. A., Kim, K. I., Ritchie, K. J. & Zhang, D-E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981 (2002).
Gong, L., Kamitani, T., Millas, S. & Yeh, E. T. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 275, 14212–14216 (2000).
Hospenthal, M. K., Freund, S. M. V. & Komander, D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat. Struct. Mol. Biol. 20, 555–565 (2013).
Castañeda, C. A., Kashyap, T. R., Nakasone, M. A., Krueger, S. & Fushman, D. Unique structural, dynamical, and functional properties of k11-linked polyubiquitin chains. Structure 21, 1168–1181 (2013).
Cook, W. J., Jeffrey, L. C., Carson, M., Chen, Z. & Pickart, C. M. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J. Biol. Chem. 267, 16467–16471 (1992).
Keusekotten, K. et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 (2013).
Thrower, J. S., Hoffman, L., Rechsteiner, M. & Pickart, C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 (2000).
Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Jinek, M. et al. RNA-programmed genome editing in human cells. elife 2, e00471 (2013).
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D. & Chin, J. W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Reyes-Turcu, F. E. et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124, 1197–1208 (2006).
Licchesi, J. D. F. et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID’s specificity for atypical ubiquitin chains. Nat. Struct. Mol. Biol. 19, 62–71 (2012).
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052 (2011).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Kirkpatrick, D. S. et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 8, 700–710 (2006).
Nishikawa, H. et al. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 (2004).
Morris, J. R. & Solomon, E. BRCA1 : BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 13, 807–817 (2004).
Wu-Baer, F., Lagrazon, K., Yuan, W. & Baer, R. The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J. Biol. Chem. 278, 34743–34746 (2003).
Lim, K. L. et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25, 2002–2009 (2005).
Durcan, T. M. et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 10.15252/embj.201489729 (2014).
Lokireddy, S. et al. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 16, 613–624 (2012).
Cheng, E. H. Y., Sheiko, T. V., Fisher, J. K., Craigen, W. J. & Korsmeyer, S. J. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science 301, 513–517 (2003).
Armstrong, L. C., Saenz, A. J. & Bornstein, P. Metaxin 1 interacts with metaxin 2, a novel related protein associated with the mammalian mitochondrial outer membrane. J. Cell. Biochem. 74, 11–22 (1999).
Yun, J. et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. elife 3, e01958 (2014).
Braschi, E., Zunino, R. & McBride, H. M. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 10, 748–754 (2009).
Hasson, S. A. et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 (2013).
Leboucher, G. P. et al. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 47, 547–557 (2012).
Crosas, B. et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127, 1401–1413 (2006).
Hanna, J. et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006).
Kirkin, V., McEwan, D. G., Novak, I. & Dikic, I. A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 (2009).
Dong, K. C. et al. Preparation of distinct ubiquitin chain reagents of high purity and yield. Structure 19, 1053–1063 (2011).
Torres, J. Z., Miller, J. J. & Jackson, P. K. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 9, 2888–2891 (2009).
Pinheiro, J., Bates, D., DebRoy, S. & Sarkar, D. (R Development Core Team) nlme: linear and nonlinear mixed effects models. R package version 3.1-101 (2011).
Acknowledgements
The authors wish to acknowledge C. Balakarski, A. Decker and W. Forrest for assistance with bioinformatic tools and statistical analyses, Y. Franke for assistance with construct design and creation and E. Dueber, J. Lill and D. Vucic for critical reading of the manuscript. PTMScan is carried out at Genentech under limited license from Cell Signaling Technologies.
Author information
Authors and Affiliations
Contributions
C.N.C., J.M.B., L.P., J.S.T., B.B. and J.E.C. were responsible for the experimental work. C.N.C., D.S.K., B.B. and J.E.C. planned the project and designed the experiments. C.N.C., J.M.B., L.P., D.S.K., B.B. and J.E.C. analysed the data. C.Y. and M.C. provided materials and reagents. C.N.C. and J.E.C. wrote the manuscript with help from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare competing financial interests. C.N.C., J.M.B., L.P., C.Y., M.C., D.S.K. and B.B. are all employees of Genentech, Inc.
Integrated supplementary information
Supplementary Figure 4 Lys6 ubiquitin chains are enriched in HEK293 and SH-SY5Y whole cell lysates and mitochondria upon CCCP treatment.
(A) SH-SY5Y Western blot confirmation of mitochondrial depolarization upon CCCP treatment over time. MFN1 levels decrease and Opa1 processing increased over the course of 4 h with 10 μM CCCP treatment in SH-SY5Y cells. (B) Base peak chromatograms of immunoaffinity-purified K-GG peptides from SH-SY5Y cells following Parkin overexpression (oe) and/or 10 μM CCCP treatment for 2 h. Chromatographic features showing ubiquitin signature peptides for Lys6, Lys11, Lys48, and Lys63-linked chains are denoted by arrows. Inset shows extracted ion chromatograms from Lys6 peptides in the treated and untreated samples. (C) SDS-PAGE gel regions excised for ubiquitin linkage analysis in Supplementary Fig. 1D. HEK293 cells stably expressing GFP-Parkin were treated with either DMSO or 20 μM CCCP for 3 h, fractionated into mitochondria, and lysed for trypsinization and subsequent ubiquitin linkage analysis. Data represent one out of three independent experiments. (D) Ubiquitin linkage analysis on 4 gel regions ranging from 5–250 kDa. Abundance (fmol) of each ubiquitin linkage is reported for DMSO (black circles) or CCCP (red circles) treatment for each gel region. Data represent one out of three experiments. (E) Representative SDS-PAGE gel showing excised regions for ubiquitin linkage analysis in Supplementary Fig. 1F. SH-SY5Y cells were treated with either DMSO or 10 μM CCCP for 3 h, fractionated into cytosolic and mitochondrial fractions, and lysed for trypsinization and subsequent ubiquitin linkage analysis. (F) Ubiquitin linkage analysis of cytosolic and mitochondrial fractions of 2 gel regions ranging from 40–100 kDa. Abundance (fmol) of each ubiquitin linkage is reported for DMSO (black circles) or CCCP (red circles) treatment for each gel region from either the cytosolic or mitochondrial fractions of two replicate experiments in SH-SY5Y cells.
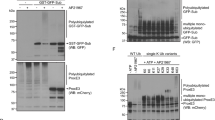
Supplementary Figure 5 Immunodepletion of GFP-Parkin confirms substrates other than Parkin carry Lys6-linked polyubiquitin chains.
(A) HEK293 cells overexpressing GFP-Parkin were treated with 10 μM CCCP or a DMSO vehicle control for 2 h. Lysates were immunodepleted with anti-GFP beads and input and flow through (FT) fractions were run on Western blot. MFN1 levels decreased and Opa1 processing increased confirming mitochondrial depolarization upon CCCP treatment. This experiment was performed once. (B) Input and flow through (FT) fractions treated with CCCP from (A) were run on SDS-PAGE and excised into 10 gel regions spanning 5–250 kDa. (C) Ubiquitin linkage analysis was used to measure the abundance of Lys6 chains (fmol) for each gel region excised (left) and Parkin peptide abundance (AUC) was measured for each gel region excised (right).
Supplementary Figure 6 USP30 is an ubiquitin specific deubiquitinase that prefers compact chain types.
(A) Michaelis–Menten plot of USP2 (blue), USP30 (orange), and USP7 (red) catalytic domain activity. Data were fit using the equation v = Vmax[S]/KM + [S]. (B) USP30’s preference for compact ubiquitin linkages is not driven by a hyper affinity for dimeric ubiquitin chain types in a biolayer interferometry experiment. Equilibrium response is shown as a function of di-ubiquitin concentration. Compact chains (Lys6, Lys11, and Lys48) are shown in colors. Greater overall signal was observed with Lys6 dimers, but individual kinetic curves displayed some non-specific adherence to the tip and fitting showed no significant (NS) decrease in dissociation constant (all KDs > 5 μM). (C) Gel of Lys6, Lys48, and Lys63 purified tetramers. The lower band of Lys6 (∗) migrates most closely to Lys63 tetramers. (D) Intact mass spectrometry of Lys6 tetramer shows a single molecular weight peak at the expected MW of tetrameric ubiquitin. (E) SDS-PAGE gel of Lys6 tetramers showing regions excised for ubiquitin linkage analysis. (F) Relative amounts of detected chain types for each excised band in (E). Lys63 make up a larger fraction of linkages in the lower band indicating this chain type as an off-target product during the enzymatic formation of K6 tetramers.
Supplementary Figure 7 Mass spectrometry validation and ubiquitin linkage analysis of USP30 knockout cell line.
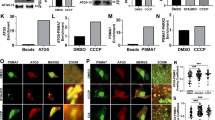
(A,B) Mass spectrometry confirmation of USP30 CRISPR knockout. Ratios of area under the curve (AUC) between USP30 knockout HEK293 cells expressing GFP-Parkin and a wild type USP30 isogenic cell line for 2 mitochondrial matrix proteins (HSPD1, ATP5B) and three independent USP30 peptides (A; see Supplementary Table 2 for raw data). Ratios are not shown for the three USP30 signature peptides since no signal was detected in the USP30 knockout cell line, as represented in (B). Error bars represent standard deviation from n = 3 independent experiments (Supplementary Table 2). (C) Representative SDS-PAGE gel from three independent experiments showing excision locations used for ubiquitin linkage analysis in Fig. 3a and Supplementary Fig. 3D. USP30 knockout (KO) or WT USP30 (WT) HEK293 cells stably expressing GFP-Parkin were treated with either DMSO or CCCP (10 μM, 3 h), enriched for mitochondria, and lysed for trypsinization and subsequent ubiquitin linkage analysis. (D) Ubiquitin linkage analysis on 4 gel regions ranging from 5–250 kDa. fmol of each ubiquitin linkage is reported for CCCP treated WT (black circles) or USP30 knockout (red circles) mitochondrial enriched fractions for each gel region. Data represent one out of two replicate experiments. (E) Representative SDS-PAGE gel showing regions excised for ubiquitin linkage analysis in Fig. 3c.
Supplementary Figure 8 USP30 deubiquitinates outer membrane mitochondrial substrates.
(A) Western blot confirmation of USP30 induction following doxycycline administration and mitochondrial depolarization upon CCCP treatment. USP30 showed strong induction, while reduced MFN1 levels and increase in Opa1 processing were observed. Representative Western blot shown from experiment 1. (B,C) Volcano plots of ubiquitinated proteins identified in Experiments 1 and 2, respectively. The fold change of ubiquitination for each protein between Combo (USP30 overexpression and CCCP) versus CCCP is denoted on the x-axis with significance reported on the y-axis. Both values were determined based on the aggregate values of individual KGG peptides using LiME analysis. Proteins with p-value < 0.01 are labelled; mitochondria-associated proteins are colored red.
Supplementary Figure 9 Atypical ubiquitin chain linkages signal for mitophagy.
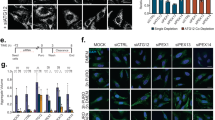
(A) Immunostaining of HeLa cells co-transfected with GFP-Parkin and mitochondrial-localized, FLAG-tagged DUBs. 24 h after transfection cells were immunostained for FLAG (green) and endogenous HSP60 (red). Each DUB co-localizes with HSP60 indicating proper mitochondrial targeting (see merge). In addition, expression levels appear uniform across all DUBs transfected. Representative cells are shown for each transfection. Scale bar, 10 μm. (B) Large field images from Fig. 5a. Solid yellow outlines highlight cells with GFP-Parkin and DUB-Flag expressing cells. Dashed yellow outlines highlight cells expressing only GFP-Parkin. Scale bar, 30 μm. (C) Immunostaining of HeLa cells co-transfected with GFP-Parkin and HA-tagged ubiquitin variants as listed in Fig. 5c. Only GFP-Parkin expressing cells are outlined. Solid lines indicate TOM20 positive staining cells; dashed lines indicate TOM20 negative staining cells. Scale bar, 20 μm. (D) Quantification of TOM20 in HeLa cells treated with CCCP (15 μM, 24 h) and transfected with either beta-Gal or HA-ubiquitin. A significant difference in TOM20 was observed with a 24 h CCCP treatment preventing analysis of individual HA-ubiquitin mutants tested in Fig. 5c. Each point represents a single experiment with a mean intensity from 25–40 cells measured; the line represents the mean from n = 5 experiments and error bars represent s.e.m. Significance is reported as one-Way ANOVA with Holm–Sidak’s multiple comparison test (∗P < 0.05).
Supplementary information
Supplementary Information
Supplementary Information (PDF 2980 kb)
Supplementary Table 1
Supplementary Information (XLSX 49 kb)
Supplementary table 2
Supplementary Information (XLSX 41 kb)
Supplementary table 3
Supplementary Information (XLSX 35 kb)
Supplementary table 4
Supplementary Information (XLS 166 kb)
Supplementary table 5
Supplementary Information (XLSX 3578 kb)
Rights and permissions
About this article
Cite this article
Cunningham, C., Baughman, J., Phu, L. et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat Cell Biol 17, 160–169 (2015). https://doi.org/10.1038/ncb3097
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3097
This article is cited by
-
Mitochondrial CISD1/Cisd accumulation blocks mitophagy and genetic or pharmacological inhibition rescues neurodegenerative phenotypes in Pink1/parkin models
Molecular Neurodegeneration (2024)
-
USP30 inhibition induces mitophagy and reduces oxidative stress in parkin-deficient human neurons
Cell Death & Disease (2024)
-
USP30 promotes the progression of breast cancer by stabilising Snail
Cancer Gene Therapy (2024)
-
The mitophagy pathway and its implications in human diseases
Signal Transduction and Targeted Therapy (2023)
-
Deubiquitinases in cancer
Nature Reviews Cancer (2023)