Abstract
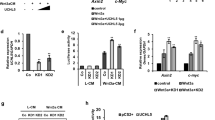
The Wnt/β-catenin signalling pathway plays essential roles in embryonic development and adult tissue homeostasis, and deregulation of this pathway has been linked to cancer. Axin is a concentration-limiting component of the β-catenin destruction complex, and its stability is regulated by tankyrase. However, the molecular mechanism by which tankyrase-dependent poly(ADP-ribosyl)ation (PARsylation) is coupled to ubiquitylation and degradation of axin remains undefined. Here, we identify RNF146, a RING-domain E3 ubiquitin ligase, as a positive regulator of Wnt signalling. RNF146 promotes Wnt signalling by mediating tankyrase-dependent degradation of axin. Mechanistically, RNF146 directly interacts with poly(ADP-ribose) through its WWE domain, and promotes degradation of PARsylated proteins. Using proteomics approaches, we have identified BLZF1 and CASC3 as further substrates targeted by tankyrase and RNF146 for degradation. Thus, identification of RNF146 as a PARsylation-directed E3 ligase establishes a molecular paradigm that links tankyrase-dependent PARsylation to ubiquitylation. RNF146-dependent protein degradation may emerge as a major mechanism by which tankyrase exerts its function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Logan, C. Y. & Nusse, R. The Wnt signalling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004).
Clevers, H. Wnt/β-catenin signalling in development and disease. Cell 127, 469–480 (2006).
Lee, E., Salic, A., Kruger, R., Heinrich, R. & Kirschner, M. W. The roles of APC and Axin derived from experimental and theoretical analysis of the Wnt pathway. PLoS. Biol. 1, E10 (2003).
Leung, J. Y. et al. Activation of AXIN2 expression by β-catenin-T cell factor. A feedback repressor pathway regulating Wnt signalling. J. Biol. Chem. 277, 21657–21665 (2002).
Willert, K., Shibamoto, S. & Nusse, R. Wnt-induced dephosphorylation of axin releases β-catenin from the axin complex. Genes Dev. 13, 1768–1773 (1999).
Huang, S. M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Hsiao, S. J. & Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92 (2008).
Gagne, J. P., Hendzel, M. J., Droit, A. & Poirier, G. G. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr. Opin. Cell Biol. 18, 145–151 (2006).
Yeh, T. Y. et al. Tankyrase recruitment to the lateral membrane in polarized epithelial cells: regulation by cell–cell contact and protein poly(ADP-ribosyl)ation. Biochem. J. 399, 415–425 (2006).
Aravind, L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275 (2001).
Chen, B. et al. Small molecule-mediated disruption of Wnt-dependent signalling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 (2009).
Sbodio, J. I. & Chi, N. W. Identification of a tankyrase-binding motif sharedby IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J. Biol. Chem. 277, 31887–31892 (2002).
Short, B. et al. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, 877–883 (2001).
Chi, N. W. & Lodish, H. F. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J. Biol. Chem. 275, 38437–38444 (2000).
Palacios, I. M., Gatfield, D., St, J. D. & Izaurralde, E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427, 753–757 (2004).
Hunter, T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 (2007).
Min, J. H. et al. Structure of an HIF-1 α-pVHL complex: hydroxyproline recognition in signalling. Science 296, 1886–1889 (2002).
Ikura, T. et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell Biol. 27, 7028–7040 (2007).
Yoshida, Y. et al. E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 (2002).
Schreiber, V., Dantzer, F., Ame, J. C. & de, M. G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 (2006).
Hassa, P. O. & Hottiger, M. O. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 13, 3046–3082 (2008).
Scovassi, A. I. The poly(ADP-ribosylation) story: a long route from Cinderella to Princess. Riv. Biol. 100, 351–360 (2007).
Karras, G. I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005).
Pleschke, J. M., Kleczkowska, H. E., Strohm, M. & Althaus, F. R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Nusse, R., van, O. A., Cox, D., Fung, Y. K. & Varmus, H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 307, 131–136 (1984).
Tsukamoto, A. S., Grosschedl, R., Guzman, R. C., Parslow, T. & Varmus, H. E. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell 55, 619–625 (1988).
Mohinta, S., Wu, H., Chaurasia, P. & Watabe, K. Wnt pathway and breast cancer. Front. Biosci. 12, 4020–4033 (2007).
Howe, L. R. & Brown, A. M. Wnt signalling and breast cancer. Cancer Biol. Ther. 3, 36–41 (2004).
Gold, B. et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl Acad. Sci. USA 105, 4340–4345 (2008).
Acknowledgements
We thank D. Patel, C. Xin, E. McWhinnie, S. Zhao, J. Murphy, Y. Mishina and J. Klekota for technical assistance and W. Shao, F. Stegmeier, J. Tallarico, T. Bouwmeester and M. Kirschner for comments and advice.
Author information
Authors and Affiliations
Contributions
Y.Z., C.M., Y.F., G.A.M., M.S., M.H., A.B., V.E.M, P.M.F., J.A.P., S-M.A.H and F.C. conceived and designed the study. Y.Z., S.L., C.M., Y.F., O.C., G.A.M., M.S., X.S. and F.C. designed and implemented experiments. Y.Z. and F.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 997 kb)
Supplementary Table 1
Supplementary Information (XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Liu, S., Mickanin, C. et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol 13, 623–629 (2011). https://doi.org/10.1038/ncb2222
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2222