Abstract
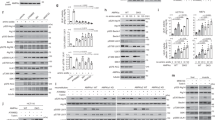
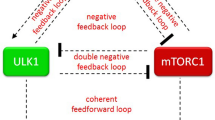
Autophagy is a process by which components of the cell are degraded to maintain essential activity and viability in response to nutrient limitation. Extensive genetic studies have shown that the yeast ATG1 kinase has an essential role in autophagy induction. Furthermore, autophagy is promoted by AMP activated protein kinase (AMPK), which is a key energy sensor and regulates cellular metabolism to maintain energy homeostasis. Conversely, autophagy is inhibited by the mammalian target of rapamycin (mTOR), a central cell-growth regulator that integrates growth factor and nutrient signals. Here we demonstrate a molecular mechanism for regulation of the mammalian autophagy-initiating kinase Ulk1, a homologue of yeast ATG1. Under glucose starvation, AMPK promotes autophagy by directly activating Ulk1 through phosphorylation of Ser 317 and Ser 777. Under nutrient sufficiency, high mTOR activity prevents Ulk1 activation by phosphorylating Ulk1 Ser 757 and disrupting the interaction between Ulk1 and AMPK. This coordinated phosphorylation is important for Ulk1 in autophagy induction. Our study has revealed a signalling mechanism for Ulk1 regulation and autophagy induction in response to nutrient signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Wang, R. C. & Levine, B. Autophagy in cellular growth control. FEBS Lett. 584, 1417–1426 (2010).
Hara, T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008).
Stipanuk, M. H. Macroautophagy and its role in nutrient homeostasis. Nutr. Rev. 67, 677–689 (2009).
Huang, J. & Klionsky, D. J. Autophagy and human disease. Cell Cycle 6, 1837–1849 (2007).
Liang, C. & Jung, J. U. Autophagy genes as tumor suppressors. Curr. Opin. Cell Biol. 22, 226–233 (2010).
Sarkar, S. & Rubinsztein, D. C. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 275, 4263–4270 (2008).
Cadwell, K., Stappenbeck, T. S. & Virgin, H. W. Role of autophagy and autophagy genes in inflammatory bowel disease. Curr. Top. Microbiol. Immunol. 335, 141–167 (2009).
Lerena, M. C., Vazquez, C. L. & Colombo, M. I. Bacterial pathogens and the autophagic response. Cell Microbiol. 12, 10–18 (2010).
Tal, M. C. & Iwasaki, A. Autophagy and innate recognition systems. Curr. Top. Microbiol. Immunol. 335, 107–121 (2009).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Xie, Z. & Klionsky, D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 (2007).
Inoue, Y. & Klionsky, D. J. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin. Cell Dev. Biol. 21, 664–670 (2010).
Mizushima, N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 (2010).
Chan, E. Y. & Tooze, S. A. Evolution of Atg1 function and regulation. Autophagy 5, 758–765 (2009).
Kamada, Y. et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513 (2000).
Kabeya, Y. et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16, 2544–2553 (2005).
Chang, Y. Y. & Neufeld, T. P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20, 2004–2014 (2009).
Chan, E. Y., Kir, S. & Tooze, S. A. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282, 25464–25474 (2007).
Young, A. R. et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 (2006).
Ganley, I. G. et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Hosokawa, N. et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979 (2009).
Jung, C. H. et al. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Sudarsanam, S. & Johnson, D. E. Functional consequences of mTOR inhibition. Curr. Opin. Drug Discov. Devel. 13, 31–40 (2010).
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M. & Kim, D. H. mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 (2010).
Chang, Y. Y. et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem. Soc. Trans. 37, 232–236 (2009).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Kamada, Y., Sekito, T. & Ohsumi, Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 279, 73–84 (2004).
Funakoshi, T., Matsuura, A., Noda, T. & Ohsumi, Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192, 207–213 (1997).
Kamada, Y. et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 30, 1049–1058 (2010).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785 (2007).
Vingtdeux, V. et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-β peptide metabolism. J. Biol. Chem. 285, 9100–9113 (2010).
Herrero-Martin, G. et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 28, 677–685 (2009).
Matsui, Y. et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 100, 914–922 (2007).
Liang, J. et al. The energy sensing LKB1–AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 9, 218–224 (2007).
Meley, D. et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 281, 34870–34879 (2006).
Inoki, K., Zhu, T. & Guan, K. L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003).
Gwinn, D. M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K. & Koike, T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteomics 5, 749–757 (2006).
Zhou, G. et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 (2001).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008).
Hawley, S. A., Gadalla, A. E., Olsen, G. S. & Hardie, D. G. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes 51, 2420–2425 (2002).
Inoki, K., Li, Y., Xu, T. & Guan, K. L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 (2003).
Zhang, Y. et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5, 578–581 (2003).
Schalm, S. S., Fingar, D. C., Sabatini, D. M. & Blenis, J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 (2003).
Inoki, K., Li, Y., Zhu, T., Wu, J. & Guan, K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002).
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Lee, J. W., Park, S., Takahashi, Y. & Wang, H. G. The association of AMPK with ULK1 regulates autophagy. PLoS One 5, e15394 (2010).
Scott, J. W., Norman, D. G., Hawley, S. A., Kontogiannis, L. & Hardie, D. G. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J. Mol. Biol. 317, 309–323 (2002).
Koren, I., Reem, E. & Kimchi, A. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093–1098 (2010).
Acknowledgements
We thank members of the Guan lab for discussions and reagents. We would especially like to thank I. Lian and C. Fang for technical assistance, and M. Farquhar, K. Kudicka and T. Meerloo for help with the electron microscopy. This work was supported by NIH grants GM51586 and GM62694 (to K.-L.G.).
Author information
Authors and Affiliations
Contributions
J.K. performed the experiments; M.K. and B.V. established the AMPK and Ulk1 knockout MEFs, respectively; J.K. and K.-L.G. designed the experiments, analysed data and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1287 kb)
Rights and permissions
About this article
Cite this article
Kim, J., Kundu, M., Viollet, B. et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141 (2011). https://doi.org/10.1038/ncb2152
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2152
This article is cited by
-
Icariin promotes bone marrow mesenchymal stem cells osteogenic differentiation via the mTOR/autophagy pathway to improve ketogenic diet-associated osteoporosis
Journal of Orthopaedic Surgery and Research (2024)
-
DEL-1: a promising treatment for AMD-associated ER stress in retinal pigment epithelial cells
Journal of Translational Medicine (2024)
-
Arginyltransferase 1 modulates p62-driven autophagy via mTORC1/AMPk signaling
Cell Communication and Signaling (2024)
-
Icariin exerts anti-tumor activity by inducing autophagy via AMPK/mTOR/ULK1 pathway in triple-negative breast cancer
Cancer Cell International (2024)
-
Multipronged regulation of autophagy and apoptosis: emerging role of TRIM proteins
Cellular & Molecular Biology Letters (2024)