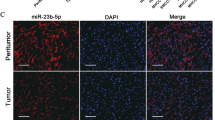
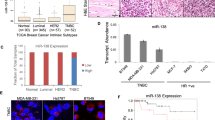
Abstract
Recurrent chromosomal aberrations are often observed in hepatocellular carcinoma (HCC), but little is known about the functional non-coding sequences, particularly microRNAs (miRNAs), at the chromosomal breakpoints in HCC. Here we show that 22 miRNAs are often amplified or deleted in HCC. MicroRNA-151 (miR-151), a frequently amplified miRNA on 8q24.3, is correlated with intrahepatic metastasis of HCC. We further show that miR-151, which is often expressed together with its host gene FAK, encoding focal adhesion kinase, significantly increases HCC cell migration and invasion in vitro and in vivo, mainly through miR-151-5p, but not through miR-151-3p. Moreover, miR-151 exerts this function by directly targeting RhoGDIA, a putative metastasis suppressor in HCC, thus leading to the activation of Rac1, Cdc42 and Rho GTPases. In addition, miR-151 can function synergistically with FAK to enhance HCC cell motility and spreading. Thus, our findings indicate that chromosome gain of miR-151 is a crucial stimulus for tumour invasion and metastasis of HCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Albertson, D. G., Collins, C., McCormick, F. & Gray, J. W. Chromosome aberrations in solid tumors. Nature Genet. 34, 369–376 (2003).
Bayani, J. et al. Genomic mechanisms and measurement of structural and numerical instability in cancer cells. Semin. Cancer Biol. 17, 5–18 (2007).
El-Serag, H. B. & Rudolph, K. L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 (2007).
Lau, S. H. & Guan, X. Y. Cytogenetic and molecular genetic alterations in hepatocellular carcinoma. Acta Pharmacol. Sin. 26, 659–665 (2005).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 5, 522–531 (2004).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nature Rev. Cancer 6, 857–866 (2006).
Esquela-Kerscher, A. & Slack, F. J. Oncomirs — microRNAs with a role in cancer. Nature Rev. Cancer 6, 259–269 (2006).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Yanaihara, N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 (2006).
Calin, G. A. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA 101, 2999–3004 (2004).
Marchio, A. et al. Distinct chromosomal abnormality pattern in primary liver cancer of non-B, non-C patients. Oncogene 19, 3733–3738 (2000).
Parada, L. A. et al. Frequent rearrangements of chromosomes 1, 7, and 8 in primary liver cancer. Genes Chromosomes Cancer 23, 26–35 (1998).
Marchio, A. et al. Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 18, 59–65 (1997).
Mihara, M. et al. Absence of mutation of the p73 gene localized at chromosome 1p36.3 in hepatocellular carcinoma. Br. J. Cancer 79, 164–167 (1999).
Hammond, C., Jeffers, L., Carr, B. I. & Simon, D. Multiple genetic alterations, 4q28, a new suppressor region, and potential gender differences in human hepatocellular carcinoma. Hepatology 29, 1479–1485 (1999).
Koshikawa, K. et al. Allelic imbalance at 1p36 in the pathogenesis of human hepatocellular carcinoma. Hepatogastroenterology 51, 186–191 (2004).
Yeh, S. H. et al. Chromosomal allelic imbalance evolving from liver cirrhosis to hepatocellular carcinoma. Gastroenterology 121, 699–709 (2001).
Fang, W., Piao, Z., Simon, D., Sheu, J. C. & Huang, S. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer 28, 269–275 (2000).
Nishimura, T. et al. Discrete breakpoint mapping and shortest region of overlap of chromosome arm 1q gain and 1p loss in human hepatocellular carcinoma detected by semiquantitative microsatellite analysis. Genes Chromosomes Cancer 42, 34–43 (2005).
Li, S. P. et al. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J. Hepatol. 34, 840–849 (2001).
Zhang, S. H., Cong, W. M., Xian, Z. H. & Wu, M. C. Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol. 11, 3034–3039 (2005).
Dykema, K. J. & Furge, K. A. Diminished transcription of chromosome arm 4q is inversely related to local spreading of hepatocellular carcinoma. Genes Chromosomes Cancer 41, 390–394 (2004).
Zhang, L. H. et al. Allelic imbalance regions on chromosomes 8p, 17p and 19p related to metastasis of hepatocellular carcinoma: comparison between matched primary and metastatic lesions in 22 patients by genome-wide microsatellite analysis. J. Cancer Res. Clin. Oncol. 129, 279–286 (2003).
Katoh, H. et al. Genetic profile of hepatocellular carcinoma revealed by array-based comparative genomic hybridization: identification of genetic indicators to predict patient outcome. J. Hepatol. 43, 863–874 (2005).
Chen, Y. J. et al. Chromosomal changes and clonality relationship between primary and recurrent hepatocellular carcinoma. Gastroenterology 119, 431–440 (2000).
Yeh, S. H. et al. Allelic loss of chromosome 4q21 approximately 23 associates with hepatitis B virus-related hepatocarcinogenesis and elevated α-fetoprotein. Hepatology 40, 847–854 (2004).
Okabe, H. et al. Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus-positive and -negative tumors. Hepatology 31, 1073–1079 (2000).
Kahng, Y. S. et al. Loss of heterozygosity of chromosome 8p and 11p in the dysplastic nodule and hepatocellular carcinoma. J. Gastroenterol. Hepatol. 18, 430–436 (2003).
Liao, C. et al. Identification of the gene for a novel liver-related putative tumor suppressor at a high-frequency loss of heterozygosity region of chromosome 8p23 in human hepatocellular carcinoma. Hepatology 32, 721–727 (2000).
Pineau, P. et al. Identification of three distinct regions of allelic deletions on the short arm of chromosome 8 in hepatocellular carcinoma. Oncogene 18, 3127–3134 (1999).
Chan, K. L., Lee, J. M., Guan, X. Y., Fan, S. T. & Ng, I. O. High-density allelotyping of chromosome 8p in hepatocellular carcinoma and clinicopathologic correlation. Cancer 94, 3179–3185 (2002).
Xu, Z., Liang, L., Wang, H., Li, T. & Zhao, M. HCRP1, a novel gene that is downregulated in hepatocellular carcinoma, encodes a growth-inhibitory protein. Biochem. Biophys. Res. Commun. 311, 1057–1066 (2003).
Piao, Z. et al. Homozygous deletions of the CDKN2 gene and loss of heterozygosity of 9p in primary hepatocellular carcinoma. Cancer Lett. 122, 201–207 (1998).
Liew, C. T. et al. Frequent allelic loss on chromosome 9 in hepatocellular carcinoma. Int. J. Cancer 81, 319–324 (1999).
Zhu, G. N. et al. Loss of heterozygosity on chromosome 10q22–10q23 and 22q11.2–22q12.1 and p53 gene in primary hepatocellular carcinoma. World J Gastroenterol 10, 1975–1978 (2004).
Ricketts, S. L., Carter, J. C. & Coleman, W. B. Identification of three 11p11.2 candidate liver tumor suppressors through analysis of known human genes. Mol Carcinog 36, 90–99 (2003).
Chow, V. T., Lim, K. M. & Lim, D. The human DENN gene: genomic organization, alternative splicing, and localization to chromosome 11p11.21–p11.22. Genome 41, 543–552 (1998).
Lin, Y. W. et al. Loss of heterozygosity at chromosome 13q in hepatocellular carcinoma: identification of three independent regions. Eur. J. Cancer 35, 1730–1734 (1999).
Wong, C. M., Lee, J. M., Lau, T. C., Fan, S. T. & Ng, I. O. Clinicopathological significance of loss of heterozygosity on chromosome 13q in hepatocellular carcinoma. Clin. Cancer Res. 8, 2266–2272 (2002).
Piao, Z., Park, C., Kim, J. J. & Kim, H. Deletion mapping of chromosome 16q in hepatocellular carcinoma. Br. J. Cancer 80, 850–854 (1999).
Wang, G. et al. Allelic loss and gain, but not genomic instability, as the major somatic mutation in primary hepatocellular carcinoma. Genes Chromosomes Cancer 31, 221–227 (2001).
Yuan, B. Z., Zhou, X., Zimonjic, D. B., Durkin, M. E. & Popescu, N. C. Amplification and overexpression of the EMS 1 oncogene, a possible prognostic marker, in human hepatocellular carcinoma. J. Mol. Diagn. 5, 48–53 (2003).
Schlaeger, C. et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology 47, 511–520 (2008).
Mitra, S. K., Hanson, D. A. & Schlaepfer, D. D. Focal adhesion kinase: in command and control of cell motility. Nature Rev. Mol. Cell Biol. 6, 56–68 (2005).
Miranda, K. C. et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 (2006).
Dovas, A. & Couchman, J. R. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390, 1–9 (2005).
von Sengbusch, A. et al. Focal adhesion kinase regulates metastatic adhesion of carcinoma cells within liver sinusoids. Am. J. Pathol. 166, 585–596 (2005).
Zhang, L. et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA 103, 9136–9141 (2006).
Thorgeirsson, S. S., Lee, J. S. & Grisham, J. W. Functional genomics of hepatocellular carcinoma. Hepatology 43, S145–S150 (2006).
McLean, G. W. et al. The role of focal-adhesion kinase in cancer — a new therapeutic opportunity. Nature Rev. Cancer 5, 505–515 (2005).
Ozsolak, F. et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 22, 3172–3183 (2008).
Corcoran, D. L. et al. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One 4, e5279 (2009).
Golubovskaya, V., Kaur, A. & Cance, W. Cloning and characterization of the promoter region of human focal adhesion kinase gene: nuclear factor κB and p53 binding sites. Biochim. Biophys. Acta 1678, 111–125 (2004).
Golubovskaya, V. M. et al. p53 regulates FAK expression in human tumor cells. Mol. Carcinog. 47, 373–382 (2008).
Loo, L. W. et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 64, 8541–8549 (2004).
Kim, S. W. et al. Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer 46, 1–9 (2007).
Saramaki, O. R., Porkka, K. P., Vessella, R. L. & Visakorpi, T. Genetic aberrations in prostate cancer by microarray analysis. Int. J. Cancer 119, 1322–1329 (2006).
Togawa, A. et al. Progressive impairment of kidneys and reproductive organs in mice lacking Rho GDIα. Oncogene 18, 5373–5380 (1999).
Ishizaki, H. et al. Defective chemokine-directed lymphocyte migration and development in the absence of Rho guanosine diphosphate-dissociation inhibitors α and β. J. Immunol. 177, 8512–8521 (2006).
Maddala, R., Reneker, L. W., Pendurthi, B. & Rao, P. V. Rho GDP dissociation inhibitor-mediated disruption of Rho GTPase activity impairs lens fiber cell migration, elongation and survival. Dev. Biol. 315, 217–231 (2008).
Blackhall, F. H. et al. Validating the prognostic value of marker genes derived from a non-small cell lung cancer microarray study. Lung Cancer 46, 197–204 (2004).
Jiang, W. G. et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin. Cancer Res. 9, 6432–6440 (2003).
DerMardirossian, C., Schnelzer, A. & Bokoch, G. M. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 15, 117–127 (2004).
Acknowledgements
We thank Didier Trono (School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland) for providing the pWPXL, psPAX2 and pMD2.G plasmids. This work was supported by grants from the Science and Technology Commission of Shanghai Municipality (07DJ14006), the Ministry of Human Resources and Social Security of China (2007-170), and the Ministry of Health of China (2008ZX10002-017).
Author information
Authors and Affiliations
Contributions
X.H., S.H., H.T. and D.W. planned the experimental design. J.D., S.H., S.W., Y.Z., L.L., M.-X.Y., C.G., J.Y., T.C., M.Y. and J.L. conducted the experiments. J.D., S.H. and X.H. analysed data. X.H., S.H., J.D., H.W. and J.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1067 kb)
Supplementary Information
Supplementary Table 1 (XLS 36 kb)
Supplementary Information
Supplementary Table 2 (XLS 68 kb)
Supplementary Information
Supplementary Table 3 (XLS 20 kb)
Supplementary Information
Supplementary Table 4 (XLS 32 kb)
Supplementary Information
Supplementary Table 5 (XLS 29 kb)
Supplementary Information
Supplementary Table 6 (XLS 26 kb)
Rights and permissions
About this article
Cite this article
Ding, J., Huang, S., Wu, S. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol 12, 390–399 (2010). https://doi.org/10.1038/ncb2039
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2039
This article is cited by
-
SDF-1 and NOTCH signaling in myogenic cell differentiation: the role of miRNA10a, 425, and 5100
Stem Cell Research & Therapy (2023)
-
The miR151 and miR5100 Transfected Bone Marrow Stromal Cells Increase Myoblast Fusion in IGFBP2 Dependent Manner
Stem Cell Reviews and Reports (2022)
-
RhoGDI1 interacts with PHLDA2, suppresses the proliferation, migration, and invasion of trophoblast cells, and participates in the pathogenesis of preeclampsia
Human Cell (2022)
-
Mutational drivers of cancer cell migration and invasion
British Journal of Cancer (2021)
-
Targeting the cytoskeleton against metastatic dissemination
Cancer and Metastasis Reviews (2021)