Abstract
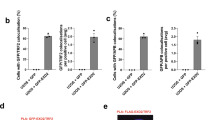
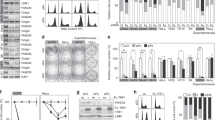
Telomerase-negative cancer cells maintain their telomeres through the alternative lengthening of telomeres (ALT) pathway1,2,3. Although a growing body of evidence demonstrates that the ALT mechanism is a post-replicative telomere recombination process, molecular details of this pathway are largely unknown. Here we demonstrate that MUS81, a DNA structure specific recombination endonuclease, has a key role in the maintenance of telomeres in human ALT cells. We find that MUS81 specifically localizes to ALT-associated promyelocytic leukaemia (PML) nuclear bodies (APBs) and associates with telomeric DNA in ALT cells, which is enriched during the G2 phase of the cell cycle. Depletion of MUS81 results in the reduction of ALT-specific telomere recombination and leads to proliferation arrest of ALT cells. In addition, the endonuclease activity of MUS81 is required for recombination-based ALT cell survival, and the interaction of MUS81 with the telomeric repeat-binding factor TRF2 regulates this enzymatic activity, thereby maintaining telomere recombination. Thus, our results suggest that MUS81 is involved in the maintenance of ALT cell survival at least in part by homologous recombination of telomeres.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Dunham, M. A., Neumann, A. A., Fasching, C. L. & Reddel, R. R. Telomere maintenance by recombination in human cells. Nature Genet. 26, 447–450 (2000).
Londono-Vallejo, J. A., Der-Sarkissian, H., Cazes, L., Bacchetti, S. & Reddel, R. R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 64, 2324–2327 (2004).
Wang, R. C., Smogorzewska, A. & de Lange, T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119, 355–368 (2004).
Cesare, A. J. & Reddel, R. R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129, 99–108 (2008).
Bailey, S. M., Brenneman, M. A. & Goodwin, E. H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 32, 3743–3751 (2004).
Yeager, T. R. et al. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59, 4175–4179 (1999).
Bryan, T. M., Englezou, A., Gupta, J., Bacchetti, S. & Reddel, R. R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248 (1995).
Muntoni, A. & Reddel, R. R. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14 Spec No. 2, R191–R196 (2005).
Jiang, W. Q., Zhong, Z. H., Henson, J. D. & Reddel, R. R. Identification of candidate alternative lengthening of telomeres genes by methionine restriction and RNA interference. Oncogene 26, 4635–4647 (2007).
Wu, G., Lee, W. H. & Chen, P. L. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 275, 30618–30622 (2000).
Grobelny, J. V., Godwin, A. K. & Broccoli, D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J. Cell Sci. 113 Pt 24, 4577–4585 (2000).
Blais, V. et al. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell 15, 552–562 (2004).
Ehmsen, K. T. & Heyer, W. D. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 36, 2182–2195 (2008).
Osman, F. & Whitby, M. C. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 6, 1004–1017 (2007).
Chen, X. B. et al. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8, 1117–1127 (2001).
Boddy, M. N. et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548 (2001).
Hollingsworth, N. M. & Brill, S. J. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18, 117–125 (2004).
Heyer, W. D. Recombination: Holliday junction resolution and crossover formation. Curr. Biol. 14, R56–R58 (2004).
Gao, H., Chen, X. B. & McGowan, C. H. Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell 14, 4826–4834 (2003).
Perrem, K., Colgin, L. M., Neumann, A. A., Yeager, T. R. & Reddel, R. R. Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21, 3862–3875 (2001).
Cerone, M. A., Londono-Vallejo, J. A. & Bacchetti, S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum. Mol. Genet. 10, 1945–1952 (2001).
de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005).
Palm, W. & de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 (2008).
Griffith, J. D. et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999).
Stansel, R. M., de Lange, T. & Griffith, J. D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20, 5532–5540 (2001).
Dendouga, N. et al. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol. Cell. Biol. 25, 7569–7579 (2005).
Yang, Q., Zheng, Y. L. & Harris, C. C. POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell. Biol. 25, 1070–1080 (2005).
Garcia-Cao, M., Gonzalo, S., Dean, D. & Blasco, M. A. A role for the Rb family of proteins in controlling telomere length. Nature Genet. 32, 415–419 (2002).
Gonzalo, S. et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature Cell Biol. 8, 416–424 (2006).
Karlseder, J., Smogorzewska, A. & de Lange, T. Senescence induced by altered telomere state, not telomere loss. Science 295, 2446–2449 (2002).
Kaliraman, V., Mullen, J. R., Fricke, W. M., Bastin-Shanower, S. A. & Brill, S. J. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730–2740 (2001).
Zhang, R. et al. BLM helicase facilitates Mus81 endonuclease activity in human cells. Cancer Res. 65, 2526–2531 (2005).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Acknowledgements
We thank R. R. Reddel for ALT cells, and C. H. McGowan for wild-type and mutant MUS81 constructs, the MUS81 antibody and Mus81 +/+ and Mus81−/− MEFs. We thank I. Hickson and J. Roti Roti for proof-reading. This work is supported in part by grants from Concern Foundation to Q.Y. and NIH CA10445/CA123232 to T.K.P. This research was also supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Contributions
S.Z. performed most of the experiments; T.X performed immunoprecipitation-western assays and I.G.S. performed Q-FISH assay; T.K.P, S.G and C.C.H analysed data and Q.Y. planned the project, designed experiments, analysed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1450 kb)
Rights and permissions
About this article
Cite this article
Zeng, S., Xiang, T., Pandita, T. et al. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol 11, 616–623 (2009). https://doi.org/10.1038/ncb1867
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1867
This article is cited by
-
Pathway choice in the alternative telomere lengthening in neoplasia is dictated by replication fork processing mediated by EXD2’s nuclease activity
Nature Communications (2023)
-
TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells
Nature Communications (2021)
-
RETRACTED ARTICLE: Identification of protein kinase inhibitors to reprogram breast cancer cells
Cell Death & Disease (2018)
-
Control of structure-specific endonucleases to maintain genome stability
Nature Reviews Molecular Cell Biology (2017)
-
Telomere recombination pathways: tales of several unhappy marriages
Current Genetics (2017)