Abstract
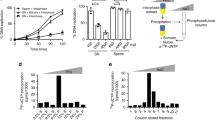
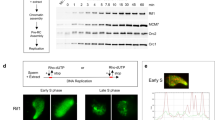
In early Xenopus development, transcription is repressed and DNA replication initiates at non-specific sites. Here, we show that a site-specific DNA replication origin can be induced in this context by the assembly of a transcription domain. Deletion of the promoter element abolishes site-specific initiation, and its relocalization to an ectopic site induces a new origin of replication. This process does not require active transcription, and specification of the origin occurs mainly through a decrease in non-specific initiation at sites distant from the promoter. Finally, chromatin immunoprecipitation experiments suggest that site-specific acetylation of histones favours the selection of the active DNA replication origin. We propose that the specification of active DNA replication origins occurs by secondary epigenetic events and that the programming of chromatin for transcription during development contributes to this selection in higher eukaryotes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
DePamphilis, M.L. Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21, 5–16 (1999).
Mechali, M. DNA replication origins: from sequence specificity to epigenetics. Nature Rev. Genet. 2, 640–645 (2001).
Gilbert, D.M. Making sense of eukaryotic DNA replication origins. Science 294, 96–100 (2001).
Marahrens, Y. & Stillman, B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–823 (1992).
Gomez, M. & Antequera, F. Organization of DNA replication origins in the fission yeast genome. EMBO J. 18, 5683–5690 (1999).
Hyrien, O., Maric, C. & Mechali, M. Transition in specification of embryonic metazoan DNA replication origins. Science 270, 994–997 (1995).
Sasaki, T., Sawado, T., Yamaguchi, M. & Shinomiya, T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19, 547–555 (1999).
Maric, C., Benard, M. & Pierron, G. Developmentally regulated usage of Physarum DNA replication origins. EMBO Rep. 4, 474–478 (2003).
Lunyak, V.V., Ezrokhi, M., Smith, H.S. & Gerbi, S.A. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol. Cell. Biol. 22, 8426–8437 (2002).
Harland, R.M. & Laskey, R.A. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell 21, 761–771 (1980).
Mechali, M. & Kearsey, S. Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 38, 55–64 (1984).
Prioleau, M.N., Buckle, R.S. & Mechali, M. Programming of a repressed but committed chromatin structure during early development. EMBO J. 14, 5073–5084 (1995).
Giacca, M. et al. Fine mapping of a replication origin of human DNA. Proc. Natl Acad. Sci. USA 91, 7119–7123 (1994).
Kitsberg, D., Selig, S., Keshet, I. & Cedar, H. Replication structure of the human β-globin gene domain. Nature 366, 588–590 (1993).
Pelizon, C., Diviacco, S., Falaschi, A. & Giacca, M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol. Cell. Biol. 16, 5358–5364 (1996).
Kobayashi, T., Rein, T. & DePamphilis, M.L. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18, 3266–3277 (1998).
Wu, J.R. & Gilbert, D.M. A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271, 1270–1272 (1996).
Rein, T., Kobayashi, T., Malott, M., Leffak, M. & DePamphilis, M.L. DNA methylation at mammalian replication origins. J. Biol. Chem. 274, 25792–25800 (1999).
Bielinsky, A.K. & Gerbi, S.A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279, 95–98 (1998).
Abdurashidova, G. et al. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287, 2023–2026 (2000).
Van der Vliet, P.C. in Concepts in DNA Replication in Eukaryotic Cells (ed. DePamphilis, M.L.) 87–118 (Cold Spring Harbor Laboratory Press, New York, 1999).
Lee, T.I. & Young, R.A. Transcription of eukaryotic protein-coding genes. Ann. Rev. Genet. 34, 77–137 (2000).
Prioleau, M.N., Huet, J., Sentenac, A. & Mechali, M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77, 439–449 (1994).
Modak, S.P., Principaud, E. & Spohr, G. Regulation of Xenopus c-myc promoter activity in oocytes and embryos. Oncogene 8, 645–654 (1993).
Schaarschmidt, D., Baltin, J., Stehle, I.M., Lipps, H.J. & Knippers, R. An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. EMBO J. 23, 191–201 (2004).
Jacob, F., Brenner, J. & Cuzin, F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 28, 329–348 (1963).
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003).
Felsenfeld, G. Chromatin as an essential part of the transcriptional mechanism. Nature 355, 219–224 (1992).
Wolffe, A.P. Chromatin: Structure and Function. (Acad. Press, San Diego, CA, 1998).
Cheng, L. & Kelly, T.J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell 59, 541–551 (1989).
Guo, Z.S. & DePamphilis, M.L. Specific transcription factors stimulate simian virus 40 and polyomavirus origins of DNA replication. Mol. Cell. Biol. 12, 2514–2524 (1992).
Li, R. & Botchan, M.R. Acidic transcription factors alleviate nucleosome-mediated repression of DNA replication of bovine papillomavirus type 1. Proc. Natl Acad. Sci. USA 91, 7051–7055 (1994).
Harvey, K.J. & Newport, J. CpG methylation of DNA restricts prereplication complex assembly in Xenopus egg extracts. Mol. Cell. Biol. 23, 6769–6779 (2003).
Workman, J.L. & Kingston, R.E. Nucleosome core displacement in vitro via a metastable transcription factor–nucleosome complex. Science 258, 1780–1784 (1992).
Cote, J., Quinn, J., Workman, J.L. & Peterson, C.L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265, 53–60 (1994).
Utley, R.T. et al. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394, 498–502 (1998).
Ryan, M.P., Stafford, G.A., Yu, L. & Morse, R.H. Artificially recruited TATA-binding protein fails to remodel chromatin and does not activate three promoters that require chromatin remodeling. Mol. Cell. Biol. 20, 5847–5857 (2000).
Vashee, S. et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17, 1894–1908 (2003).
Prasanth, S.G., Prasanth, K.V. & Stillman, B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297, 1026–1031 (2002).
Romanowski, P., Madine, M.A., Rowles, A., Blow, J.J. & Laskey, R.A. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 6, 1416–1425 (1996).
Carpenter, P.B., Mueller, P.R. & Dunphy, W.G. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature 379, 357–360 (1996).
Royzman, I., Austin, R.J., Bosco, G., Bell, S.P. & Orr-Weaver, T.L. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13, 827–840 (1999).
Kong, D., Coleman, T.R. & DePamphilis, M.L. Xenopus origin recognition complex (ORC) initiates DNA replication preferentially at sequences targeted by Schizosaccharomyces pombe ORC. EMBO J. 22, 3441–3450 (2003).
Lu, Z.H., Sittman, D.B., Romanowski, P. & Leno, G.H. Histone H1 reduces the frequency of initiation in Xenopus egg extract by limiting the assembly of prereplication complexes on sperm chromatin. Mol. Biol. Cell 9, 1163–1176 (1998).
Bouvet, P., Dimitrov, S. & Wolffe, A.P. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 8, 1147–1159 (1994).
Giacca, M., Pelizon, C. & Falaschi, A. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods 13, 301–312 (1997).
Acknowledgements
We thank D. Fisher for critical reading of this manuscript. This study has been supported by grants from the CNRS, Human Frontier Science Program, Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale (FRM) and the Ligue Nationale Contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Fig. S1, Fig. S2, Fig. S3,,Fig. S4, Fig. S5 and supplementary methods (PDF 1172 kb)
Rights and permissions
About this article
Cite this article
Danis, E., Brodolin, K., Menut, S. et al. Specification of a DNA replication origin by a transcription complex. Nat Cell Biol 6, 721–730 (2004). https://doi.org/10.1038/ncb1149
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1149
This article is cited by
-
A predictable conserved DNA base composition signature defines human core DNA replication origins
Nature Communications (2020)
-
The gastrula transition reorganizes replication-origin selection in Caenorhabditis elegans
Nature Structural & Molecular Biology (2017)
-
DNA replication origin activation in space and time
Nature Reviews Molecular Cell Biology (2015)
-
The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage
Nature Communications (2015)
-
Epigenetic landscape for initiation of DNA replication
Chromosoma (2014)