Abstract
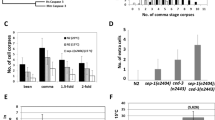
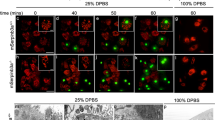
Engulfment of apoptotic cells in Caenorhabditis elegans is controlled by two partially redundant pathways. Mutations in genes in one of these pathways, defined by the genes ced-2, ced-5 and ced-10, result in defects both in the engulfment of dying cells and in the migrations of the two distal tip cells of the developing gonad. Here we find that ced-2 and ced-10 encode proteins similar to the human adaptor protein CrkII and the human GTPase Rac, respectively. Together with the previous observation that ced-5 encodes a protein similar to human DOCK180, our findings define a signalling pathway that controls phagocytosis and cell migration. We provide evidence that CED-2 and CED-10 function in engulfing rather than dying cells to control the phagocytosis of cell corpses, that CED-2 and CED-5 physically interact, and that ced-10 probably functions downstream of ced-2 and ced-5. We propose that CED-2/CrkII and CED-5/DOCK180 function to activate CED-10/Rac in a GTPase signalling pathway that controls the polarized extension of cell surfaces.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fadok, V. A. & Henson, P. M. Apoptosis: getting rid of the bodies. Curr. Biol. 8, R693–R695 (1998).
Metzstein, M. M., Stanfield, G. M. & Horvitz, H. R. Genetics of programmed cell death in C. elegans: Past, present and future. Trends Genet. 14, 410–416 (1998).
Adams, J. M. & Cory, S. The Bcl-2 family: arbiters of cell survival. Science 281, 322–326 (1998).
Hedgecock, E. M., Sulston, J. E. & Thomson, J. N. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220, 1277–1279 (1983).
Ellis, R. E., Jacobson, D. M. & Horvitz, H. R. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129, 79–94 (1991).
Wu, Y. C. & Horvitz, H. R. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392, 501–504 (1998).
Liu, Q. A. & Hengartner, M. O. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93, 961–972 (1998).
Wu, Y. C. & Horvitz, H. R. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93, 951–960 (1998).
Hasegawa, H. et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell Biol. 16, 1770–1776 (1996).
Erickson, M., Galletta, B. J. & Abmayr, S. M. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure, and cytoskeletal organization. J. Cell Biol. 138, 589–603 (1997).
Kimble, J. & Hirsh, D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70, 396–417 (1979).
The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 (1998).
Matsuda, M. et al. Two species of human CRK cDNA encode proteins with distinct biological activities. Mol. Cell Biol. 12, 3482–3489 (1992).
Mayer, B. J., Hamaguchi, M. & Hanafusa, H. A novel viral oncogene with structural similarity to phospholipase C. Nature 332, 272–275 (1988).
Kiyokawa, E., Hashimoto, Y., Kurata, T., Sugimura, H. & Matsuda, M. Evidence that DOCK180 up-regulates signals from the CrkII-p130(Cas) complex. J. Biol. Chem. 273, 24479–24484 (1998).
Klemke, R. L. et al. CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol. 140, 961–972 (1998).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Imaizumi, T. et al. Mutant mice lacking crk-II caused by the gene trap insertional mutagenesis: crk-II is not essential for embryonic development. Biochem. Biophys. Res. Commun. 266, 569–574 (1999).
ten Hoeve, J., Morris, C., Heisterkamp, N. & Groffen, J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene 8, 2469–2474 (1993).
Alfonso, A., Grundahl, K., Duerr, J. S., Han, H. P. & Rand, J. B. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261, 617–619 (1993).
Li, W., Herman, R. K. & Shaw, J. E. Analysis of the Caenorhabditis elegans axonal guidance and outgrowth gene unc-33. Genetics 132, 675–689 (1992).
Chen, W., Lim, H. H. & Lim, L. A new member of the ras superfamily, the rac1 homologue from Caenorhabditis elegans. J. Biol. Chem. 268, 320–324 (1993).
Van Aelst, L. & D"Souza-Schorey, C. Rho GTPases and signaling networks. Genes Dev. 11, 2295–2322 (1997).
Caron, E. & Hall, A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282, 1717–1721 (1998).
Massol, P., Montcourrier, P., Guillemot, J. C. & Chavrier, P. Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17, 6219–6229 (1998).
Kiyokawa, E. et al. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 12, 3331–3336 (1998).
Nolan, K. M. et al. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 12, 3337–3342 (1998).
Li, L. & Cohen, S. N. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85, 319–329 (1996).
Spieth, J., Brooke, G., Kuersten, S., Lea, K. & Blumenthal, T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73, 521–532 (1993).
Zhang, F. L. & Casey, P. J. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 (1996).
Reiss, Y., Stradley, S. J., Gierasch, L. M., Brown, M. S. & Goldstein, J. L. Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc. Natl Acad. Sci. USA 88, 732–736 (1991).
Bourne, H. R., Sanders, D. A. & McCormick, F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 (1991).
Sugihara, K. et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 17, 3427–3433 (1998).
Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. & Hengartner, M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011–1022 (1999).
Lauffenburger, D. A. & Horwitz, A. F. Cell migration: a physically integrated molecular process. Cell 84, 359–369 (1996).
Platt, N., da Silva, R. P. & Gordon, S. Recognizing death: the phagocytosis of apoptotic cells. Trends Cell Biol. 8, 365–372 (1998).
Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Bloom, L. & Horvitz, H. R. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl Acad. Sci. USA 94, 3414–3419 (1997).
Krause, M. & Hirsh, D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell 49, 753–761 (1987).
Acknowledgements
We thank B. Hersh and R. Ranganathan for comments concerning this manuscript, and Y. Kohara for providing cDNA clones. P.W.R. was supported by a National Science Foundation Fellowship and an NIH training grant. H.R.H. is an Investigator of the Howard Hughes Medical Institute.
Correspondence and requests for materials should be addressed to H.R.H. The nucleotide sequences of ced-2 and ced-10 have been deposited at GenBank under accession numbers AF226866 and AF226867, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reddien, P., Horvitz, H. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol 2, 131–136 (2000). https://doi.org/10.1038/35004000
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/35004000
This article is cited by
-
The cisd gene family regulates physiological germline apoptosis through ced-13 and the canonical cell death pathway in Caenorhabditis elegans
Cell Death & Differentiation (2019)
-
Interpreting an apoptotic corpse as anti-inflammatory involves a chloride sensing pathway
Nature Cell Biology (2019)
-
Mutations in dock1 disrupt early Schwann cell development
Neural Development (2018)
-
Engulfing cells promote neuronal regeneration and remove neuronal debris through distinct biochemical functions of CED-1
Nature Communications (2018)
-
The Small GTPase RAC1/CED-10 Is Essential in Maintaining Dopaminergic Neuron Function and Survival Against α-Synuclein-Induced Toxicity
Molecular Neurobiology (2018)