Abstract
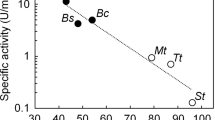
Thermostable enzymes combine catalytic specificity with the toughness required to withstand industrial reaction conditions1,2. Stabilized enzymes also provide robust starting points for evolutionary improvement of other protein properties3. We recently created a library4 of at least 2,300 new active chimeras of the biotechnologically important5,6,7,8,9 cytochrome P450 enzymes. Here we show that a chimera's thermostability can be predicted from the additive contributions of its sequence fragments. Based on these predictions, we constructed a family of 44 novel thermostable P450s with half-lives of inactivation at 57 °C up to 108 times that of the most stable parent. Although they differ by as many as 99 amino acids from any known P450, the stable sequences are catalytically active. Among the novel functions they exhibit is the ability to produce drug metabolites. This chimeric P450 family provides a unique ensemble for biotechnological applications and for studying sequence-stability-function relationships.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
12 December 2007
In the print version of this article and the version initially published online, the sentence starting with “The most thermostable 450 P450 chimera” contains an extra “450”. The correct sentence should start as follows “The most thermostable P450 chimera”.
References
Niehaus, F., Bertoldo, C., Kahler, M. & Antranikian, G. Extremophiles as a source of novel enzymes for industrial application. Appl. Microbiol. Biotechnol. 51, 711–729 (1999).
Zeikus, J.G., Vieille, C. & Savchenko, A. Thermozymes: biotechnology and structure-function relationships. Extremophiles 2, 179–183 (1998).
Bloom, J.D., Labthavikul, S.T., Otey, C.R. & Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 103, 5869–5874 (2006).
Otey, C.R. et al. Structure-guided recombination creates an artificial family of cytochromes P450. PLoS Biol. 4, e112 (2006).
Guengerich, F.P. Cytochrome P450 enzymes in the generation of commercial products. Nat. Rev. Drug Discov. 1, 359–366 (2002).
Urlacher, V.B. & Eiben, S. Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol. 24, 324–330 (2006).
Landwehr, M. et al. Enantioselective alpha-hydroxylation of 2-arylacetic acid derivatives and buspirone catalyzed by engineered cytochrome P450BM-3. J. Am. Chem. Soc. 128, 6058–6059 (2006).
Otey, C.R., Bandara, G., Lalonde, J., Takahashi, K. & Arnold, F.H. Preparation of human metabolites of propranolol using laboratory-evolved bacterial cytochromes P450. Biotechnol. Bioeng. 93, 494–499 (2006).
van Vugt-Lussenburg, B.M., Damsten, M.C., Maasdijk, D.M., Vermeulen, N.P. & Commandeur, J.N. Heterotropic and homotropic cooperativity by a drug-metabolising mutant of cytochrome P450BM3. Biochem. Biophys. Res. Commun. 346, 810–818 (2006).
Landwehr, M., Carbone, M., Otey, C.R., Li, Y. & Arnold, F.H. Diversification of catalytic function in a synthetic family of chimeric cytochrome P450s. Chem. Biol. 4, 269–278 (2007).
Dietterich, T.G. Approximate statistical tests for comparing supervised classification learning algorithms. Neural Comput. 10, 1895–1923 (1998).
Fox, R. et al. Optimizing the search algorithm for protein engineering by directed evolution. Protein Eng. 16, 589–597 (2003).
Amin, N. et al. Construction of stabilized proteins by combinatorial consensus mutagenesis. Protein Eng. Des. Sel. 17, 787–793 (2004).
Lehmann, M. et al. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15, 403–411 (2002).
Steipe, B., Schiller, B., Pluckthun, A. & Steinbacher, S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 240, 188–192 (1994).
Johannes, T.W., Woodyer, R.D. & Zhao, H.M. Directed evolution of a thermostable phosphite dehydrogenase for NAD(P)H regeneration. Appl. Environ. Microbiol. 71, 5728–5734 (2005).
Abernethy, D.R., Wainer, I.W., Longstreth, J.A. & Andrawis, N.S. Stereoselective verapamil disposition and dynamics in aging during racemic verapamil administration. J. Pharmacol. Exp. Ther. 266, 904–911 (1993).
Busse, D., Cosme, J., Beaune, P., Kroemer, H.K. & Eichelbaum, M. Cytochromes of the P450 2C subfamily are the major enzymes involved in the O-demethylation of verapamil in humans. Naunyn Schmiedebergs Arch. Pharmacol. 353, 116–121 (1995).
Meuldermans, W., Hendrickx, J., Lauwers, W., Hurkmans, R., Swysen, E. & Heykants, J. Excretion and biotransformation of astemizole in rats, guinea-pigs, dogs, and man. Drug Dev. Res. 8, 37–51 (1986).
Heykants, J., Peer, A.V., Woestenborghs, R., Jageneaur, A. & Bussche, G.V. Dose-proportionality, bioavailability, and steady-state kinetics of astemizole in man. Drug Dev. Res. 8, 71–78 (1986).
Wells, J.A. Additivity of mutational effects in proteins. Biochemistry 29, 8509–8517 (1990).
Meyer, M.M., Hochrein, L. & Arnold, F.H. Structure-guided SCHEMA recombination of distantly related beta-lactamases. Protein Eng. Des. Sel. 19, 563–570 (2006).
Acknowledgements
The authors thank Christopher R. Otey and Marco Landwehr for their assistance. This work was supported in part by National Institutes of Health Grant R01 GM068664-01, Army Research Office Contract DAAD19-03-0004, Eli Lilly & Co. and a Howard Hughes Medical Institute predoctoral fellowship (to J.D.B.).
Author information
Authors and Affiliations
Contributions
Y.L., A.M.S. and F.H.A. designed the research; Y.L. and A.M.S. performed the research; Y.L., D.A.D., A.M.S., C.D.S. and J.D.B. contributed to the analytical tools; Y.L., D.A.D., A.M.S., C.D.S., J.D.B. and F.H.A. analyzed data; Y.L., D.A.D., A.M.S., C.D.S. and F.H.A. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The thermostable P450s described have been licensed for commercialization. Remuneration is already fixed and will not be affected by this paper. F.S.A. is in talks to serve as consultant to licensor.
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–6 and Supplementary Tables 1–7 (PDF 148 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Drummond, D., Sawayama, A. et al. A diverse family of thermostable cytochrome P450s created by recombination of stabilizing fragments. Nat Biotechnol 25, 1051–1056 (2007). https://doi.org/10.1038/nbt1333
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1333
This article is cited by
-
Self-driving laboratories to autonomously navigate the protein fitness landscape
Nature Chemical Engineering (2024)
-
Searching for protein variants with desired properties using deep generative models
BMC Bioinformatics (2023)
-
Consequences of Genetic Recombination on Protein Folding Stability
Journal of Molecular Evolution (2023)
-
Hydropathy and Conformational Similarity-Based Distributed Representation of Protein Sequences for Properties Prediction
SN Computer Science (2022)
-
CRISPR technologies and the search for the PAM-free nuclease
Nature Communications (2021)