Abstract
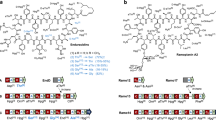
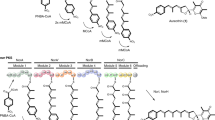
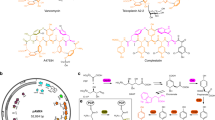
Molecular engineering approaches to producing new antibiotics have been in development for about 25 years. Advances in cloning and analysis of antibiotic gene clusters, engineering biosynthetic pathways in Escherichia coli, transfer of engineered pathways from E. coli into Streptomyces expression hosts, and stable maintenance and expression of cloned genes have streamlined the process in recent years. Advances in understanding mechanisms and substrate specificities during assembly by polyketide synthases, nonribosomal peptide synthetases, glycosyltransferases and other enzymes have made molecular engineering design and outcomes more predictable. Complex molecular scaffolds not amenable to synthesis by medicinal chemistry (for example, vancomycin (Vancocin), daptomycin (Cubicin) and erythromycin) are now tractable by molecular engineering. Medicinal chemistry can further embellish the properties of engineered antibiotics, making the two disciplines complementary.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Newman, D.J., Cragg, G.M. & Snader, K.M. Natural products as sources of new drugs over the period 1981–2002. J. Nat. Prod. 66, 1022–1037 (2003).
Alekshun, M.N. New advances in antibiotic development and discovery. Expert Opin. Investig. Drugs 14, 117–134 (2005).
Baltz, R.H. Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall. SIM News 55, 186–196 (2005).
Baltz, R.H. Combinatorial biosynthesis of novel antibiotics and other secondary metabolites in actinomycetes. SIM News 56, 148–158 (2006).
McDaniel, R., Ebert-Khosla, S., Hopwood, D.A. & Khosla, C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature 375, 549–554 (1995).
Katz, L. Manipulation of modular polyketide synthases. Chem. Rev. 97, 2557–2576 (1997).
McDaniel, R. et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc. Natl. Acad. Sci. USA 96, 1846–1851 (1999).
Hutchinson, C.R. & McDaniel, R. Combinatorial biosynthesis in microorganisms as a route to new antimicrobial, antitumor and neuroregerative drugs. Curr. Opin. Investig. Drugs 2, 1681–1690 (2001).
Gust, B. et al. λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54, 107–128 (2004).
Vetcher, L. et al. Rapid engineering of the geldanamycin biosynthetic pathway by Red/ET recombination and gene complementation. Appl. Environ. Microbiol. 71, 1829–1835 (2005).
Baltz, R.H., Miao, V. & Wrigley, S.K. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 22, 717–741 (2005).
Baltz, R.H., Brian, P., Miao, V. & Wrigley, S.K. Combinatorial biosynthesis of lipopeptide antibiotics in Streptomyces roseosporus. J. Ind. Microbiol. Biotechnol. 33, 66–74 (2006).
Baltz, R.H. Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol. 6, 76–83 (1998).
Sanchez, C., Méndez, C. & Salas, J.A. Engineering biosynthetic pathways to generate antitumor indolecarbazole derivatives. J. Ind. Microbiol. Biotechnol. 33, 560–568 (2006).
Eustaquio, A.S. et al. Production of 8′-halogenated and 8′-unsubstituted novobiocin derivatives in genetically engineered Streptomyces coelicolor. Chem. Biol. 11, 1561–1572 (2004).
Penn, J. et al. Heterologous production of daptomycin in Streptomyces lividans. J. Ind. Microbiol. Biotechnol. 33, 121–128 (2006).
Solenberg, P.J. et al. Production of hybrid glycopeptide antibiotics in vitro and in Streptomyces toyocaensis. Chem. Biol. 4, 195–202 (1997).
Reeves, C.D. et al. Production of hybrid 16-membered macrolides by expressing combinations of polyketide synthase genes in engineered Streptomyces fradiae. Chem. Biol. 11, 1465–1472 (2004).
Bierman, M. et al. Plasmid cloning vechors for conjugal transfer from Escherichia coli to Streptomyces ssp. Gene 116, 43–49 (1992).
Miao, V. et al. Genetic engineering in Streptomyces roseosporus to produce hybrid lipopeptide antibiotics. Chem. Biol. 13, 269–276 (2006).
Lautru, S. & Challis, G.L. Substrate recognition by nonribosomal peptide synthetase multi-enzymes. Microbiology 150, 1629–1636 (2004).
Sieber, S.A. & Marahiel, M.A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005).
Grünewald, J. & Marahiel, M.A. Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol. Mol. Biol. Rev. 70, 121–146 (2006).
Hahn, M. & Stachelhaus, T. Selective interaction between nonribosomal peptide synthetases is facilitated by short communication-mediating domains. Proc. Natl. Acad. Sci. USA 101, 15585–15590 (2004).
Hahn, M. & Stachelhaus, T. Harnessing the potential of communication-mediating domains for biocombinatorial synthesis of nonribosomal peptides. Proc. Natl. Acad. Sci. USA 103, 275–280 (2006).
Pace, J.L. & Yang, G. Glycopeptides: update on as old successful antibiotic class. Biochem. Pharmacol. 71, 968–980 (2006).
Oberthur, M. et al. A systematic investigation of the synthetic utility of glycopeptide glycosyltransferases. J. Am. Chem. Soc. 127, 10747–10752 (2005).
Kahne, D., Leimkuhler, C., Lu, W. & Walsh, C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 105, 425–448 (2005).
Donadio, S., Sosio, M., Stegmann, E., Weber, T. & Wohlleben, W. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol. Genet. Genomics 274, 40–50 (2005).
Puk, O. et al. Biosynthesis of chloro-β-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186, 6093–6100 (2004).
Süssmuth, R.D. & Wohlleben, W. The biosynthesis of glycopeptide antibiotics—a model for complex, non-ribosomally synthesized, peptidic secondary metabolites. Appl. Microbiol. Biotechnol. 63, 344–350 (2004).
Bischoff, D. et al. The biosynthesis of vancomycin-type antibiotics – a model for oxidative side-chain cross-linking by oxygenases coupled to the action of peptide synthetase. ChemBioChem 6, 267–272 (2005).
Walsh, C., Freel Meyers, C.L. & Losey, H.C. Antibiotic glycosyltransferases: antibiotic maturation and prospects for reprogramming. J. Med. Chem. 46, 3425–3436 (2003).
Losey, H.C. et al. Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem. Biol. 9, 1305–1314 (2002).
Yang, J., Fu, X., Liao, J., Liu, L. & Thorson, J.S. Structure-based engineering of E. coli galactokinase as a first step toward in vivo glycorandomization. Chem. Biol. 12, 657–664 (2005).
Langenhan, J.M., Griffith, B.R. & Thorson, J.S. Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification. J. Nat. Prod. 68, 1696–1711 (2005).
Zhang, C. et al. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science 313, 1291–1294 (2006).
Kruger, R.G. et al. Tailoring of glycopeptide scaffolds by the acyltransferase from the teicoplanin and A-40,926 biosynthetic operons. Chem. Biol. 12, 131–140 (2005).
Bister, B. et al. Bromobalhimycin and chlorobromobalhimycins – illuminating the potential of halogenases in glycopeptide antibiotic biosynthesis. ChemBioChem 4, 658–662 (2003).
Weist, S. et al. Fluorobalhimycin - a new chapter in glycopeptide antibiotic research. Angew. Chem. Int. Edn. Engl. 41, 3383–3385 (2002).
Weist, S. et al. Mutasynthesis of glycopeptide antibiotics: variations of vancomycin's AB-ring amino acid 3,5-dihydroxyphenylglycine. J. Am. Chem. Soc. 126, 5942–5943 (2004).
Nguyen, K. et al. Combinatorial biosynthesis of novel antibiotics related to daptomycin. Proc. Natl. Acad. Sci. USA 103, 17462–17467 (2006).
Coëffet-Le Gal, M.-F., Thurson, L., Rich, P., Miao, V. & Baltz, R.H. Complementation of daptomycin dptA and dptD deletion mutations in trans and production of hybrid lipopeptide antibiotics. Microbiology 152, 2993–3001 (2006).
Nguyen, K.T. et al. Identification of a glutamic acid 3-methyltransferase gene by functional analysis of an accessory gene locus important for daptomycin biosynthesis in Streptomyces roseosporus. Mol. Microbiol. 61, 1294–1307 (2006).
Hojati, Z. et al. Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem. Biol. 9, 1175–1187 (2002).
Miao, V. et al. The lipopeptide antibiotic A54145 biosynthetic gene cluster from Streptomyces fradiae. J. Ind. Microbiol. Biotechnol. 33, 129–140 (2006).
Grünewald, J., Sieber, S.A., Mahlert, C., Linne, U. & Marahiel, M.A. Synthesis and derivatization of daptomycin: a chemoenzymatic route to acidic lipopeptide antibiotics. J. Am. Chem. Soc. 126, 17025–17031 (2004).
Kopp, F., Grünewald, J., Mahlert, C. & Marahiel, M.A. Chemoenzymatic design of acidic lipopeptide hybrids: new insights into the structure-activity relationship of daptomycin and A54145. Biochemistry 45, 10474–10481 (2006).
McDaniel, R., Welch, M. & Hutchinson, C.R. Genetic approaches to polyketide antibiotics.1. Chem. Rev. 105, 543–558 (2005).
Weissman, K.J. & Leadlay, P.F. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 3, 925–936 (2005).
Reid, R. et al. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42, 72–79 (2003).
Baerga-Ortiz, A. et al. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem. Biol. 13, 277–285 (2006).
O'Hare, H.M., Baerga-Ortiz, A., Popovic, B., Spencer, J.B. & Leadlay, P.F. High-throughput mutagenesis to evaluate models of stereochemical control in ketoreductase domains from the erythromycin polyketide synthase. Chem. Biol. 13, 287–296 (2006).
Menzella, H.G. et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 23, 1171–1176 (2005).
Chandran, S.S., Menzella, H.G., Carney, J.R. & Santi, D.V. Activating hybrid modular interfaces in synthetic polyketide synthases by cassette replacement of ketosynthase domains. Chem. Biol. 13, 469–474 (2006).
Sherman, D. The lego-ization of polyketide biosynthesis. Nat. Biotechnol. 23, 1083–1084 (2005).
Menzella, H.G. et al. Redesign, synthesis and functional expression of the 6-deoxyerythronolide B polyketide synthase gene cluster. J. Ind. Microbiol. Biotechnol. 33, 22–28 (2006).
Peiru, S., Menzella, H.G., Rodriquez, E., Carney, J. & Gramajo, H. Production of the potent antibacterial polyketide erythromycin C in Escherichia coli. Appl. Environ. Microbiol. 71, 2539–2547 (2005).
Borisova, S.A. et al. Substrate specificity of the macrolide-glycosylating enzyme pair DesVII/DesVIII: opportunities, limitations, and mechanistic hypothesis. Angew. Chem. Int. Edn. Engl. 45, 2748–2753 (2006).
Galm, U., Dessoy, A., Schmidt, J., Wessjohann, L.A. & Heide, L. In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic, and synthetic approach. Chem. Biol. 11, 173–183 (2004).
Eustaquio, A.S. et al. Heterologous expression of novobiocin and chlorobiocin biosynthetic gene clusters. Appl. Environ. Microbiol. 71, 2452–2459 (2005).
Li, S.-M. & Heide, L. New aminocoumarin antibiotics from genetically engineered Streptomyces strains. Curr. Med. Chem. 12, 419–427 (2005).
Flatman, R.H., Eustaquio, A., Li, S.-M., Heide, L. & Maxwell, A. Structure-activity relationships of aminocoumarin-type gyrase and topoisomerase IV inhibitors obtained by combinatorial biosynthesis. Antimicrob. Agents Chemother. 50, 1136–1142 (2006).
Waldron, C. et al. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8, 487–499 (2001).
Hahn, D.R. et al. Butenyl-spinosyns, a natural example of genetic engineering of antibiotic biosynthetic genes. J. Ind. Microbiol. Biotechnol. 33, 94–104 (2006).
Sheehan, L.S. et al. Engineering of the spinosyn PKS cluster to generate novel active spinosyn analogs. J. Nat. Prod. (in the press).
Salas, A.P. et al. Deciphering the late steps in the biosynthesis of the anti-tumor indolecarbazole staurosporine: sugar donor substrate flexibility of the StaG glycosyltransferase. Mol. Microbiol. 58, 17–27 (2005).
Sanchez, C. et al. Combinatorial biosynthesis of antitumor indolocarbazole compounds. Proc. Natl. Acad. Sci. USA 102, 461–466 (2005).
Sosio, M. & Donadio, S. Understanding and manipulating glycopeptide pathways: the example of the dalbavancin precursor A40926. J. Ind. Microbiol. Biotechnol. 33, 569–576 (2006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Baltz, R. Molecular engineering approaches to peptide, polyketide and other antibiotics. Nat Biotechnol 24, 1533–1540 (2006). https://doi.org/10.1038/nbt1265
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1265
This article is cited by
-
Microbiological and molecular insights on rare Actinobacteria harboring bioactive prospective
Bulletin of the National Research Centre (2020)
-
Combined available nitrogen resources enhanced erythromycin production and preliminary exploration of metabolic flux analysis under nitrogen perturbations
Bioprocess and Biosystems Engineering (2019)
-
Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities
Journal of Industrial Microbiology and Biotechnology (2019)
-
Module and individual domain deletions of NRPS to produce plipastatin derivatives in Bacillus subtilis
Microbial Cell Factories (2018)
-
Blocking the flow of propionate into TCA cycle through a mutB knockout leads to a significant increase of erythromycin production by an industrial strain of Saccharopolyspora erythraea
Bioprocess and Biosystems Engineering (2017)