Abstract
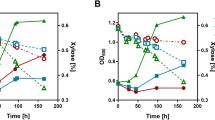
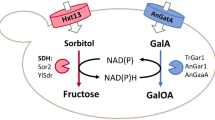
Increasing the flux through central carbon metabolism is difficult because of rigidity in regulatory structures, at both the genetic and the enzymatic levels. Here we describe metabolic engineering of a regulatory network to obtain a balanced increase in the activity of all the enzymes in the pathway, and ultimately, increasing metabolic flux through the pathway of interest. By manipulating the GAL gene regulatory network of Saccharomyces cerevisiae, which is a tightly regulated system, we produced prototroph mutant strains, which increased the flux through the galactose utilization pathway by eliminating three known negative regulators of the GAL system: Gal6, Gal80, and Mig1. This led to a 41% increase in flux through the galactose utilization pathway compared with the wild-type strain. This is of significant interest within the field of biotechnology since galactose is present in many industrial media. The improved galactose consumption of the gal mutants did not favor biomass formation, but rather caused excessive respiro-fermentative metabolism, with the ethanol production rate increasing linearly with glycolytic flux.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bailey, J.E. Lessons from metabolic engineering for functional genomics and drug discovery . Nat. Biotechnol. 17, 616– 618 (1999).
Cameron, D.C. & Tong, I.T. Cellular and metabolic engineering: an overview. Appl. Biochem. Biotechnol. 38, 105–140 (1993).
Ostergaard, S., Olsson, L. & Nielsen, J. Metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 64, 34–50 (2000).
Schaaff, I., Heinisch, J. & Zimmermann, F.K. Overproduction of glycolytic enzymes in yeast. Yeast 5, 285–290 ( 1989).
Acerenza, L. & Kacser, H. Enzyme kinetics and metabolic control. Biochem. J. 269, 697– 707 (1990).
Niederberger, P., Prasad, R., Miozzari, G. & Kacser, H. A strategy for increasing an in vivo flux by genetic manipulations. Biochem. J. 287, 473–479 (1992).
Small, R. & Kacser, H. Responses of metabolic systems to large changes in enzyme activities and effectors. J. Biochem. 213, 613–624 ( 1993).
Johnston, M. & Carlson, M. In The molecular and cellular biology of the yeast Saccharomyces: Gene expression. (eds Jones, E.W., Pringel, J.R. & Broach, J.) 193– 281 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1992).
Zheng, W., Xu, H.E. & Johnston, S.A. The cysteine–peptidase bleomycin hydrolase is a member of the galactose regulon in yeast. J. Biol. Chem. 272, 30350–30355 (1997).
Wiselogel, A., Tyson, S. & Johnson, D. In Handbook on bioethanol: production and utilization. (ed. Wyman, C.E.) 105–118 (Taylor & Francis, Washington, DC; 1996).
Ostergaard, S., Roca, C., Rønnow, B., Nielsen, J. & Olsson, L. Physiological characterisation in aerobic batch cultivations of Saccharomyces cerevisiae strains harboring the MEL1 gene. Biotechnol. Bioeng. 68, 252–259 ( 2000).
Igarashi, M., Segawa, T., Nogi, Y., Suzuki, Y. & Fukasawa, T. Autogenous regulation of the Saccharomyces cerevisiae regulatory gene GAL80. Mol. Gen. Genet. 207, 273–279 (1987).
Adam, A.C., Rubio-Texeira, M. & Polaina, J. Induced expression of bacterial β-glucosidase activity in Saccharomyces. Yeast 11, 395– 406 (1995).
Martegani, E., Brambilla, L., Porro, D., Ranzi, B.M. & Alberghina, L. Alteration of cell population structure due to cell lysis in Saccharomyces cerevisiae cells overexpressing the GAL4 gene. Yeast 9, 575–582 (1993).
Gill, G. & Ptashne, M. Negative effect of the transcriptional activator GAL4. Nature 334, 721–724 (1988).
Güldener, U., Heck, S., Fiedler, T., Beinhauer, J. & Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 (1996).
Wach, A., Brachat, A., Pöhlmann, R. & Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10, 1793–1808 (1994).
Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330 (1993).
Hoffman, C.S. & Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267– 272 (1987).
Sambrook, J., Fritsch, E.F. & Maniatis, T.M. Molecular cloning: a laboratory manual. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1989 ).
Johnston, M. & Dover, J. Mutational analysis of the gal4-encoded transcriptional regulatory protein of Saccharomyces cerevisiae . Genetics 102, 63– 74 (1988).
Gietz, D., St. Jean, A., Woods, R.A. & Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425 ( 1992).
Rose, M.D., Winston, F. & Hieter, P. Methods in yeast genetics: a laboratory course manual . (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1990).
Leuther, K.K. & Johnston, S.A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science 256, 1333–1335 (1992).
Wu, Y., Reece, R.J. & Ptashne, M. Quantitation of putative activator–target affinities predicts transcriptional activating potentials. EMBO J. 15, 3951–3963 (1996).
Suzuki-Fujimoto, T. et al. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol. Cell. Biol. 16, 2504–2508 (1996).
Yano, K. & Fukasawa, T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94, 1721–1726 ( 1997).
Keleher, C.A., Redd, M.J., Schultz, J., Carlson, M. & Johnson, A.D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68, 709–719 (1992).
Nehlin, J.O., Carlberg, M. & Ronne, H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10, 3373–3377 (1991).
Treitel, M.A. & Carlson, M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. USA 92, 3132–3136 (1995).
Acknowledgements
The research on galactose metabolism and glucose repression at the Center for Process Biotechnology has been financially supported by the Danish Programme for Food Technology II (project 2409) as well as by the European Commission Framework IV “Cell Factory” (contract BIO-CT95-0107). Peter Kötter, Goethe Universität Frankfurt, is acknowledged for providing the Δmig1Δgal80 mutant strains (SO3 and SO4).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ostergaard, S., Olsson, L., Johnston, M. et al. Increasing galactose consumption by Saccharomyces cerevisiae through metabolic engineering of the GAL gene regulatory network. Nat Biotechnol 18, 1283–1286 (2000). https://doi.org/10.1038/82400
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/82400
This article is cited by
-
Ethanol production from the red seaweed Eucheuma denticulatum and recovery of 5-hydroxymethylfurfural
Biomass Conversion and Biorefinery (2023)
-
The Metabolism of Respiring Carbon Sources by Dekkera bruxellensis and Its Relation with the Production of Acetate
Applied Biochemistry and Biotechnology (2023)
-
Uncoupling glucose sensing from GAL metabolism for heterologous lactose fermentation in Saccharomyces cerevisiae
Biotechnology Letters (2021)
-
Enhancement of Galactose Uptake from Kappaphycus alvarezii Using Saccharomyces cerevisiae through Deletion of Negative Regulators of GAL Genes
Applied Biochemistry and Biotechnology (2021)
-
Implications of agar and agarase in industrial applications of sustainable marine biomass
Applied Microbiology and Biotechnology (2020)