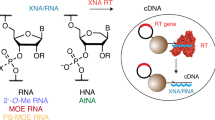
Abstract
Modified RNA and DNA molecules have novel properties that their natural counterparts do not possess, such as better resistance to degradation in cells and improved pharmacokinetic behavior1,2. In particular, modifications at the 2′-OH of ribose are important for enhancing the stability of RNA3,4. Unfortunately, it is difficult to enzymatically synthesize modified nucleic acids of any substantial length because natural polymerases incorporate modified nucleotides inefficiently. Previously, we reported an activity-based method for selecting functional T7 RNA polymerase variants based on the ability of a T7 RNA polymerase to reproduce itself5. Here, we have modified the original procedure to identify polymerases that can efficiently incorporate multiple modified nucleotides at the 2′ position of the ribose. Most important, our method allows the selection of polymerases that have good processivities and can be combined to simultaneously incorporate several different modified nucleotides in a transcript.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Opalinska, J.B. & Gewirtz, A.M. Nucleic-acid therapeutics: basic principles and recent applications. Nat. Rev. Drug Discov. 1, 503–514 (2002).
Sullenger, B.A. & Gilboa, E. Emerging clinical applications of RNA. Nature 418, 252–258 (2002).
Pieken, W.A., Olsen, D.B., Benseler, F., Aurup, H. & Eckstein, F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science 253, 314–317 (1991).
Kurreck, J. Antisense technologies Improvement through novel chemical modifications. Eur. J. Biochem. 270, 1628–1644 (2003).
Chelliserrykattil, J., Cai, G. & Ellington, A.D. A combined in vitro/in vivo selection for polymerases with novel promoter specificities. BMC Biotechnol. 1, 13 (2001).
Patel, P.H. & Loeb, L.A. DNA polymerase active site is highly mutable: evolutionary consequences. Proc. Natl. Acad. Sci. USA 97, 5095–5100 (2000).
Ghadessy, F.J., Ong, J.L. & Holliger, P. Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad. Sci. USA 98, 4552–4557 (2001).
Patel, P.H. & Loeb, L.A. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem. 275, 40266–40272 (2000).
Xia, G. et al. Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl. Acad. Sci. USA 99, 6597–6602 (2002).
Fa, M., Radeghieri, A., Henry, A.A. & Romesberg, F.E. Expanding the substrate repertoire of a DNA polymerase by directed evolution. J. Am. Chem. Soc. 126, 1748–1754 (2004).
Roychowdhury, A., Illangkoon, H., Hendrickson, C.L. & Benner, S.A. 2′-deoxycytidines carrying amino and thiol functionality: synthesis and incorporation by vent (exo(-)) polymerase. Org. Lett. 6, 489–492 (2004).
Hutter, D. & Benner, S.A. Expanding the genetic alphabet: non-epimerizing nucleoside with the pyDDA hydrogen-bonding pattern. J. Org. Chem. 68, 9839–9842 (2003).
Mitsui, T., Kimoto, M., Sato, A., Yokoyama, S. & Hirao, I. An unnatural hydrophobic base, 4-propynylpyrrole-2-carbaldehyde, as an efficient pairing partner of 9-methylimidazo[(4,5)-b]pyridine. Bioorg. Med. Chem. Lett. 13, 4515–4518 (2003).
Switzer, C.Y., Moroney, S.E. & Benner, S.A. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry 32, 10489–10496 (1993).
Hirao, I., Mitsui, T., Kimoto, M., Harada, Y. & Yokoyama, S. An unnatural base pair for efficient incorporation of nucleotide analogs into RNAs. Nucleic Acids Res. Suppl. 3, 215–216 (2003).
Sismour, A.M. et al. PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. Nucleic Acids Res. 32, 728–735 (2004).
Padilla, R. & Sousa, R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 30, e138 (2002).
Padilla, R. & Sousa, R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutant T7 RNA polymerase (RNAP). Nucleic Acids Res. 27, 1561–1563 (1999).
Huang, Y., Eckstein, F., Padilla, R. & Sousa, R. Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry 36, 8231–8242 (1997).
Brieba, L.G. & Sousa, R. Roles of histidine 784 and tyrosine 639 in ribose discrimination by T7 RNA polymerase. Biochemistry 39, 919–923 (2000).
Yin, Y.W. & Steitz, T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298, 1387–1395 (2002).
Temiakov, D. et al. Structural basis for substrate selection by T7 RNA polymerase. Cell 116, 381–391 (2004).
Sousa, R. & Padilla, R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 14, 4609–4621 (1995).
Braasch, D.A. et al. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42, 7967–7975 (2003).
Rong, M., Durbin, R.K. & McAllister, W.T. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 273, 10253–10260 (1998).
delCardayre, S.B. & Raines, R.T. Structural determinants of enzymatic processivity. Biochemistry 33, 6031–6037 (1994).
Jansen, B. & Zangemeister-Wittke, U. Antisense therapy for cancer–the time of truth. Lancet Oncol. 3, 672–683 (2002).
Majlessi, M., Nelson, N.C. & Becker, M.M. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 26, 2224–2229 (1998).
Lesnik, E.A. & Freier, S.M. What affects the effect of 2′-alkoxy modifications? 1. Stabilization effect of 2′-methoxy substitutions in uniformly modified DNA oligonucleotides. Biochemistry 37, 6991–6997 (1998).
Chelliserrykattil, J. & Ellington, A.D. in Methods Mol Biol, Directed enzyme evolution vol. 230 (eds. Arnold, F.H. & Georgiou, G.) 27–43, (Humana Press, Totowa, New Jersey, 2003).
Acknowledgements
This work was supported by a grant from the Office of Naval Research and by the Robert A. Welch Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Chelliserrykattil, J., Ellington, A. Evolution of a T7 RNA polymerase variant that transcribes 2′-O-methyl RNA. Nat Biotechnol 22, 1155–1160 (2004). https://doi.org/10.1038/nbt1001
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1001
This article is cited by
-
A two-residue nascent-strand steric gate controls synthesis of 2′-O-methyl- and 2′-O-(2-methoxyethyl)-RNA
Nature Chemistry (2023)
-
High-throughput iSpinach fluorescent aptamer-based real-time monitoring of in vitro transcription
Bioresources and Bioprocessing (2022)
-
Total RNA Synthesis and its Covalent Labeling Innovation
Topics in Current Chemistry (2022)
-
Modified nucleic acids: replication, evolution, and next-generation therapeutics
BMC Biology (2020)
-
Evolution of thermophilic DNA polymerases for the recognition and amplification of C2ʹ-modified DNA
Nature Chemistry (2016)