Abstract
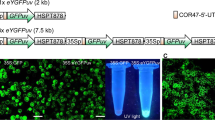
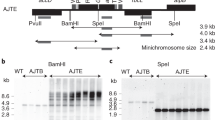
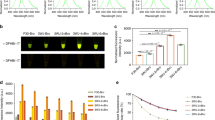
Plastid transformation in higher plants is accomplished through a gradual process, during which all the 300–10,000 plastid genome copies are uniformly altered. Antibiotic resistance genes incorporated in the plastid genome facilitate maintenance of transplastomes during this process. Given the high number of plastid genome copies in a cell, transformation unavoidably yields chimeric tissues, which requires the identification of transplastomic cells in order to regenerate plants. In the chimeric tissue, however, antibiotic resistance is not cell autonomous: transplastomic and wild-type sectors both have a resistant phenotype because of phenotypic masking by the transgenic cells. We report a system of marker genes for plastid transformation, termed FLARE-S, which is obtained by translationally fusing aminoglycoside 3"-adenyltransferase with the Aequorea victoria green fluorescent protein. 3"-adenyltransferase (FLARE-S) confers resistance to both spectinomycin and streptomycin. The utility of FLARE-S is shown by tracking segregation of individual transformed and wild-type plastids in tobacco and rice plants after bombardment with FLARE-S vector DNA and selection for spectinomycin and streptomycin resistance, respectively. This method facilitates the extension of plastid transformation to nongreen plastids in embryogenic cells of cereal crops.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Maliga, P. Towards plastid transformation in flowering plants. Trends Biotechnol. 11, 101–107 ( 1993).
Daniell, H., Datta, R., Varma, S., Gray, S. & Lee, S.B. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 16, 345– 348 (1998).
Zoubenko, O.V., Allison, L.A., Svab, Z. & Maliga, P. Efficient targeting of foreign genes into the tobacco plastid genome. Nuceic Acids Res. 22, 3819–3824 ( 1994).
Svab, Z., Hajdukiewicz, P. & Maliga, P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 87, 8526– 8530 (1990).
Svab, Z. & Maliga, P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90, 913–917 (1993).
Golds, T., Maliga, P. & Koop, H.U. Stable plastid transformation in PEG-treated protoplasts of Nicotiana tabaccum. Biotechnology 11, 95–97 (1993).
Koop, H.U. et al. Integration of foreign sequences into the tobacco plastome via PEG-mediated protoplast transformation. Planta 199, 193–101 (1996).
O'Neill, C., Horvath, G.V., Horvath, E., Dix, P.J. & Medgyesy, P. Chloroplast transformation in plants: polyethylene glycol (PEG) treatment of protoplasts is an alternative to biolistic delivery system. Plant J. 3, 729– 738 (1993).
Carrer, H., Hockenberry, T.N., Svab, Z. & Maliga, P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 241, 49– 56. (1993).
Moll, B., Posby, L. & Maliga, P. Streptomycin and lincomycin resistance are selective plastid markers in cultured Nicotiana cells. Mol. Gen. Genet. 221, 245–250 (1990).
Prasher, D.C., Eckenrode, V.K., Ward, W.W., Predergast, F.G. & Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111, 229–233 (1992).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W. & Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Heim, R., Prasher, D.C. & Tsien, R.Y. Wavelength mutations and posttranslational autooxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91, 12501–12504 (1994).
Prasher, D.C. Using GFP to see the light. Trends Genet. 11, 320–323 (1995).
Cubitt, A.B. et al. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20, 448– 455 (1995).
Misteli, T. & Spector, D.L. Applications of the green fluorescent protein in cell biology and biotechnology. Nat. Biotechnol. 15, 961–964 (1997).
Pang, S-Z. et al. An improved green fluorescent protein gene as avital marker in plants. Plant Physiol. 112, 893– 900 (1996).
Reichel, C. et al. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc. Natl. Acad. Sci. USA 93, 5888–5893 (1996).
Rouwendal., G.J.A., Mendes, O., Wolbert, E.J.H. & de Boer, A.D. Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage. Plant Mol. Biol. 33, 989–999 (1997).
Haseloff, J., Siemering, K.R., Prasher, D.C. & Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122– 2127 (1997).
Davis, S.J. & Vierstra, R.D. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528 ( 1998).
Sheen, J., Hwang, S., Niwa, Y., Kobayashi, H. & Galbraith, D.W. Green fluorescent protein as a new vital marker in plant cells. Plant J. 8, 777– 784 (1995).
Chiu, W-L. et al. Engineered gfp as a vital reporter in plants. Curr. Biol. 6, 325–330 ( 1996).
Köhler, R.H., Cao, J., Zipfel, W.R., Webb, W.W. & Hanson, M.R. Exchange of protein molecules through connections between higher plant plastids. Science 276, 2039–2042 (1997).
Baulcombe, D.C. Chapman, S. & Cruz, S.S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7, 1045– 1053 (1995).
Epel, B.L., Padgett, H.S., Heinlein, M. & Beachy, R. Plant virus movement protein dynamics probed with GFP-protein fusion. Gene 173, 75–79 ( 1996).
Hibberd, J.M., Linley, P.J., Khan, M.S. & Gray, J.C. Transient expression of green fluorescent protein in various plastid types following micro-projectile bombardment. Plant J. 16, 627– 632 (1998).
Fromm, H., Edelman, M., Aviv, D. & Galun, E. The molecular basis of rRNA-dependent spectinomycin resistance in Nicotiana chloroplasts. EMBO J. 11, 3233–3237 (1987).
Crameri, A., Whitehorn, E.A., Tate, E. & Stemmer, W.P.C. Improved green fluorescent protein by molecular evolution by DNA shuffling. Nat. Biotechnol. 14, 315– 319 (1996).
Maliga, P. Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci. 3, 4–6 (1998).
Bendich, A.J. Why do chloroplasts and mitochondria contain so many copies of their genome? Bio-essays 6, 279–282 (1987).
Sikdar, S.R., Serino, G., Chaudhuri, S. & Maliga, P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 18, 20–24 ( 1998).
Chinault, A.C. et al. Characterization of transferable plasmids for Shigella flexneri 2a that confer resistance trimethoprim, streptomycin and sulfonamides. Plasmid 15, 119–131 1986.
Staub, J.M. & Maliga, P. Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12, 601–606 (1993).
Hiratsuka, J. et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217, 185–194 (1989).
Kolodziej, P.A. & Young, R.A. Epitope tagging and protein surveillance. Methods Enzymol. 194, 508–519 (1991).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497 (1962).
Tompson, J.A., Abdullah, R. & Cocking, E.C. Protoplast culture of rice using media solidified with agarose. Plant Science 47, 123– 133 (1986).
Muller, A.J. & Grafe, R. Isolation and characterization of cell lines of Nicotiana tabacum lacking nitrate reductase. Mol. Gen. Genet. 161, 67–76 (1978).
Zhang, W. & Wu, R. Efficient regeneration of transgenic plants from rice protoplasts and correctly regulated expression of the foreign gene in the plants. Theor. Appl. Genet. 76, 835–840 (1988).
Mettler, I.J. A simple and rapid method for minipreparation of DNA from tissue-cultured plant cells. Plant Mol. Biol. Rep. 5, 346 –349 (1987).
Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature 227, 680–685 (1970).
Acknowledgements
We thank Hiroshi Kuroda for providing plasmids pHK10 and pHK34, Peter Hajdukiewicz for plasmids containing TpsbA and engineered gfp, and Millie Georgiadis for discussions concerning the design of fusion proteins. This research was supported by the Rockefeller Foundation Rice Biotechnology Program through a Postdoctoral Fellowship Award to M.S.K. and a Research Grant to P.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khan, M., Maliga, P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol 17, 910–915 (1999). https://doi.org/10.1038/12907
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/12907
This article is cited by
-
Gene editing with an oxathiapiprolin resistance selection marker reveals that PuLLP, a loricrin-like protein, is required for oospore development in Pythium ultimum
Phytopathology Research (2023)
-
Engineering of insecticidal hybrid gene into potato chloroplast genome exhibits promising control of Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae)
Transgenic Research (2023)
-
Assessing phytotoxicity of microplastics on aquatic plants using fluorescent microplastics
Environmental Science and Pollution Research (2023)
-
Engineering the plastid and mitochondrial genomes of flowering plants
Nature Plants (2022)
-
Advances in plastid transformation for metabolic engineering in higher plants
aBIOTECH (2022)