Abstract
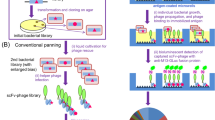
A bacteriophage λ surface expression system, λfoo, was used for epitope mapping of human galectin-3. We constructed random epitope and peptide libraries and compared their efficiencies in the mapping. The galectin-3 cDNA was randomly digested by DNase I to make random epitope libraries. The libraries were screened by affinity selection using a microtiter plate coated with monoclonal antibodies. Direct DNA sequencing of the selected clones defined two distinct epitope sites consisting of nine and 11 amino-acid residues. Affinity selection of random peptide libraries recovered a number of sequences that were similar to each other but distinct from the galectin-3 sequence. These results demonstrate that a single affinity selection of epitope libraries with antibodies is able to define an epitope determinant as small as nine residues long and is more efficient in epitope mapping than random peptide libraries.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Smith, G.P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228: 1315–1317.
Wells, J.A. and Lowman, H.B. 1992. Rapid evolution of peptide and protein binding properties in vitro. Curr. Opin. Struct. Biol. 2: 597–604.
Lu, Z., Murray, K.S., Cleave, V.V., LaVallie, E.R., Stahl, M.L., and McCoy, J.M. 1995. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: a system designed for exploring protein- protein interactions. Bio/Technology 13: 366–372.
Kasahara, N., Dozy, A.M., and Kan, Y.W. 1994. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science 266: 1373–1376.
Maruyama, I.N., Maruyama, H.I., and Brenner, S. 1994. λfoo: a λ phage vector for the expression of foreign proteins. Proc. Natl. Acad. Sci. USA 91: 8273–8277.
Sternberg, N. and Hoess, R.H. 1995. Display of peptides and proteins on the surface of bacteriophge λ. Proc. Natl. Acad. Sci. USA 92: 1609–1613.
Dunn, I.S. 1995. Assembly of functional bacteriophage lambda virions incorporating C-terminal peptide or protein fusions with the major tail protein. J. Mol. Biol. 248: 497–506.
Mikawa, Y.G., Maruyama, I.N., and Brenner, S. 1996. Surface display of proteins on bacteriophage λ heads. J. Mol. Biol. 262: 21–30.
Stanley, K.K. and Herz, J. 1987. Topological mapping of complement component C9 by recombinant DNA techniques suggests a novel mechanism for its insertion into target membranes. EMBO J. 6: 1951–1957.
Lenstra, J.A., Kusters, J.G. and van der Zeijst, B.A.M. 1990. Mapping of viral epitopes with prokaryotic expression products. Arch. Virol. 110: 1–24.
Lane, D.P. and Stephen, C.W. 1993. Epitope mapping using bacteriophage peptide libraries. Curr. Opin. Immunol. 5: 268–271.
Cortese, R., Felici, F., Galfre, G., Luzzago, A., Monaci, P., and Nicosia, A. 1994. Epitope discovery using peptide libraries displayed on phage. Trends in Biotechnology 12: 262–267.
Lam, K.S., Salmon, S.E., Hersc, E.M., Hruby, V.J., Kazmierski, W.M., and Knapp, R.J. 1991. A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354: 82–84.
Hughes, R.C. 1994. Mac-2: a versatile galactose-binding protein of mammalian tissues. Glycobiology 4: 5–12.
Robertson, M.W., Albrandt, K., Keller, D., and Liu, F.-T. 1990. Human IgE-bind-ing protein: a soluble lectin exhibiting a highly conserved interspecies sequence and differential recognition of IgE glycoforms. Biochemisty 29: 8093–8100.
Woo, H.-J., Shaw, L.M., Messier, J.M., and Mercuric, A.M. 1990. The major non-integrin laminin binding protein of macrophage is identical to carbohydrate binding protein 35 (Mac-2). J. Biol. Chem. 265: 7097–7099.
Sparrow, C.P., Leffler, H., and Barondes, S.H. 1987. Multiple soluble beta-galactoside-binding lectins from human lung. J. Biol. Chem. 262: 7383–7390.
Wang, J.L., Werner, E.A., Laing, J.G., and Patterson, R.J. 1992. Nuclear and cytoplasmic localization of a lectin-ribonucleoprotein complex. Biochem. Soc. Trans. 20: 269–274.
Liu, F.-T., Hsu, D.K., Zuberi, R.I., Hill, P.N., Shenhav, A., Kuwabara, I. et al. 1996. Modulation of functional properties of galectin-3 by monoclonal antibodies binding to the non-lectin domains. Biochemistry 36: 6073–6079.
Wang, L.-F., Plessis, D.H.D., White, J.R., Hyatt, A.D., and Eaton, B.T. 1995. Use of a gene-targeted phage display random epitope library to map an antigenie determinant on the bluetongue virus outer capsid protein VP5. J. Immunol. Method 178: 1–12.
Petersen, G., Song, D., Hugle-Dorr, B., Oldenburg, I., and Bautz, E.K.F. 1995. Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol. Gen. Genet. 249: 425–431.
Van Zonneveld, A.J. Van den Berg, B.M.M., Van Meijer, M., and Pannekoek, H. 1995. Identification of functional interaction sites on proteins using bacteriophage-displayed random epitope libraries. Gene 167: 49–52.
Maruyama, I.N., Mikawa, Y.G., and Maruyama, H.I. 1995. A model for transmembrane signalling by the aspartate receptor based on random-cassette mutagenesis and site-directed disulfide cross-linking. J. Mol. Biol. 253: 530–546.
Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: a laboratory manual, 2 ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
Yanisch-Perron, C., Vieira, J., and Messing, J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kuwabara, I., Maruyama, H., Mikawa, Y. et al. Efficient epitope mapping by bacteriophage λ surface display. Nat Biotechnol 15, 74–78 (1997). https://doi.org/10.1038/nbt0197-74
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0197-74