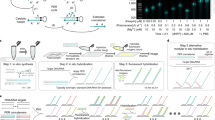
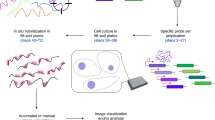
Abstract
Methods for detecting single nucleic acids in cell and tissues, such as fluorescence in situ hybridization (FISH), are limited by relatively low signal intensity and nonspecific probe binding. Here we present click-amplifying FISH (clampFISH), a method for fluorescence detection of nucleic acids that achieves high specificity and high-gain (>400-fold) signal amplification. ClampFISH probes form a 'C' configuration upon hybridization to the sequence of interest in a double helical manner. The ends of the probes are ligated together using bio-orthogonal click chemistry, effectively locking the probes around the target. Iterative rounds of hybridization and click amplify the fluorescence intensity. We show that clampFISH enables the detection of RNA species with low-magnification microscopy and in RNA-based flow cytometry. Additionally, we show that the modular design of clampFISH probes allows multiplexing of RNA and DNA detection, that the locking mechanism prevents probe detachment in expansion microscopy, and that clampFISH can be applied in tissue samples.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Itzkovitz, S. & van Oudenaarden, A. Validating transcripts with probes and imaging technology. Nat. Methods 8 (Suppl.), S12–S19 (2011).
Chen, C.H. et al. Specific sorting of single bacterial cells with microfabricated fluorescence-activated cell sorting and tyramide signal amplification fluorescence in situ hybridization. Anal. Chem. 83, 7269–7275 (2011).
Lu, J. & Tsourkas, A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 37, e100 (2009).
Banér, J., Nilsson, M., Mendel-Hartvig, M. & Landegren, U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res. 26, 5073–5078 (1998).
Larsson, C., Grundberg, I., Söderberg, O. & Nilsson, M. In situ detection and genotyping of individual mRNA molecules. Nat. Methods 7, 395–397 (2010).
Ali, M.M. et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 43, 3324–3341 (2014).
Lohman, G.J.S., Zhang, Y., Zhelkovsky, A.M., Cantor, E.J. & Evans, T.C. Jr. Efficient DNA ligation in DNA-RNA hybrid helices by Chlorella virus DNA ligase. Nucleic Acids Res. 42, 1831–1844 (2014).
Lagunavicius, A. et al. Novel application of Phi29 DNA polymerase: RNA detection and analysis in vitro and in situ by target RNA-primed RCA. RNA 15, 765–771 (2009).
Dirks, R.M. & Pierce, N.A. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA 101, 15275–15278 (2004).
Choi, H.M.T., Beck, V.A. & Pierce, N.A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014).
Shah, S. et al. Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).
Lau, J.Y. et al. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet 341, 1501–1504 (1993).
Kern, D. et al. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 34, 3196–3202 (1996).
Battich, N., Stoeger, T. & Pelkmans, L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 10, 1127–1133 (2013).
Shah, S. et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016).
Nilsson, M. et al. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265, 2085–2088 (1994).
Jin, J., Vaud, S., Zhelkovsky, A.M., Posfai, J. & McReynolds, L.A. Sensitive and specific miRNA detection method using SplintR ligase. Nucleic Acids Res. 44, e116 (2016).
Besanceney-Webler, C. et al. Increasing the efficacy of bioorthogonal click reactions for bioconjugation: a comparative study. Angew. Chem. Int. Edn. Engl. 50, 8051–8056 (2011).
Padovan-Merhar, O. et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol. Cell 58, 339–352 (2015).
Kerjaschki, D., Sharkey, D.J. & Farquhar, M.G. Identification and characterization of podocalyxin--the major sialoprotein of the renal glomerular epithelial cell. J. Cell Biol. 98, 1591–1596 (1984).
Horrillo, A., Porras, G., Ayuso, M.S. & González-Manchón, C. Loss of endothelial barrier integrity in mice with conditional ablation of podocalyxin (Podxl) in endothelial cells. Eur. J. Cell Biol. 95, 265–276 (2016).
Klemm, S. et al. Transcriptional profiling of cells sorted by RNA abundance. Nat. Methods 11, 549–551 (2014).
Bushkin, Y. et al. Profiling T cell activation using single-molecule fluorescence in situ hybridization and flow cytometry. J. Immunol. 194, 836–841 (2015).
Chen, F., Tillberg, P.W. & Boyden, E.S. Expansion microscopy. Science 347, 543–548 (2015).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Pelliccia, F., Gaddini, L., Limongi, M.Z. & Rocchi, A. Visualizing human 5S rDNA. Chromosome Res. 5, 205–207 (1997).
Mellis, I.A., Gupte, R., Raj, A. & Rouhanifard, S.H. Visualizing adenosine-to-inosine RNA editing in single mammalian cells. Nat. Methods 14, 801–804 10.1038/nmeth.4332 (2017).
Acknowledgements
We thank F. Tuluc from the CHOP flow cytometry core facility for discussions and assistance with flow cytometry. We also thank J. Peterson for his early contributions and the many bioRxiv readers who reached out with feedback and suggestions. S.H.R. acknowledges support from NIH 1F32GM120929-01A1; I.A.M. acknowledges support from NIH F30 NS100595; O.S. acknowledges support from the Human Frontier Science Program LT000919/2016-l; C.L.J. acknowledges support from NIH 5T32DK007780-19; and A.R. from NIH 4DN U01 HL129998, NIH Center for Photogenomics RM1 HG007743, the Chan Zuckerberg Initiative and HCA Pilot Project 174285, NSF CAREER 1350601 and NIH R33 EB019767.
Author information
Authors and Affiliations
Contributions
S.H.R. and A.R. designed the study and wrote the manuscript. I.A.M. performed statistical analysis. S.H.R., M.D., S.B., C.L.J., I.D., O.S., B.E., E.T., A.C., A.S. and J.A.S. designed and performed the experiments.
Corresponding author
Ethics declarations
Competing interests
A.R. receives royalties related to Stellaris RNA FISH probes.
Integrated supplementary information
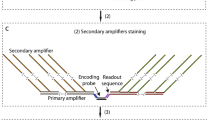
Supplementary Figure 1 Generation of ClampFISH probes.
(a) Diagram of individual pieces of each clampFISH probe. (b) Linear diagram of clampFISH probe ligation scheme. (c) ClampFISH probe ligation protocol. (d) 15% TBE-UREA gel showing separation of individual pieces, ligation product, and product after purification. (e) Varying number of landing pads on primary probes. Detected with MM2 secondary labeled in Cy5.
Supplementary Figure 2 Sensitivity and selectivity assessment.
(a) Fraction of clampFISH spots that colocalize with single molecule RNA FISH spots (blue) and spots that do not colocalize with single molecule RNA FISH spots (red) as the threshold for spot calling is relaxed. (b) (blue) Fraction of all clampFISH spots that colocalize with single molecule RNA FISH spots as the threshold is relaxed, (green) fraction of all single molecule RNA FISH spots that colocalize with clampFISH spots. Note: Single-molecule spot localization is difficult at very high fluorescence levels, making stringent spot localization difficult for bright clampFISH spots. This may contribute to an apparent (false) loss of colocalization with single molecule RNA FISH spots, whose location is easier to computationally estimate.
Supplementary Figure 3 Optimizing [formamide] in hybridization buffer.
(a) Density plots showing single spot intensity distributions across 3 different formamide concentrations in hybridization buffer. WM983b-GFP cells were probed for GFP mRNA using clampFISH probes out to round 2 (green) and WM983b cells were also stained for GFP mRNA using GFP targeting clampFISH probes (gray). (b) Mean spot counts per cell were assessed in WM983b cells (gray) and WM983b-GFP cells (green) after 2 rounds of amplification.
Supplementary Figure 6 HIST1H4E clampFISH and smFISH with high- and low-magnification scans.
HeLa cells were probed for HIST1H4E expression using clampFISH probes in Cy5 at round 8 (left) and smFISH probes in Alexa 594 (right). These are representative images from a 60X and 20X scan.
Supplementary Figure 7 Enlarged image of PODXL mRNA clampFISH on kidney tissue.
Enlarged center panel from Figure 2 showing nonspecific binding of clampFISH probes on mouse kidney tissues.
Supplementary Figure 8 Blocking reagents reduce nonspecific probe binding for flow cytometry.
MDA-MB 231 cells expressing GFP were mixed with MDA-MB 231 cells not expressing GFP at 50%. The mixed cell population was subsequently stained with clampFISH probes targeting GFP mRNA and the separation was assessed with the addition of different blocking reagents in the hybridization buffer.
Supplementary Figure 9 Biological replicate of clampFISH flow cytometry experiment.
Biological replicate of flow cytometry clampFISH from Figure 2d.
Supplementary Figure 10 Expansion clampFISH.
(a) Expansion clampFISH workflow. (b) (left) Expansion clampFISH samples were assessed for spot intensity and compared to Expansion smFISH samples targeting the same RNA (GFP). (right) Density plots showing single spot intensity distributions across all cells using expansion clampFISH vs. expansion smFISH. (c) Mean counts per cell for each experimental condition. Left represents replicate 1 and right represents replicate 2. Note that the fully expanded cells are larger than the imaging field, and the values reported are from partial cells in all experimental conditions.
Supplementary Figure 11 Assessment of bleedthrough for multiplexing.
(a) Multiplexing probe hybridization scheme. (b) Multiplexing ClampFISH to Round 7 bleedthrough. Cells were stained with clampFISH probes to round 7 individually and assessed using the same exposure times from the multiplexing experiment in Figure 3 for bleedthrough.
Supplementary Figure 12 Alternative amplification schemes.
(a) Comparison of 3 nucleic acid based amplification schemes. Pink star represents fluor. (b) Fluorescent micrographs of GFP targeting probes using each amplification method using 100X magnification. Top images are from WM983b-GFP cells and bottom images are from WM983b cells. We fixed contrast parameters independently for each method and used those for all samples of a given method; however, contrast parameters varied between methods. Images of rounds 6 and 12 of clampFISH are from Figure 1 using a lower exposure time and have been normalized to round 6 clampFISH from the current experiment. (c) Fluorescent micrographs acquired using 20X magnification on GFP positive cells (top) and GFP negative cells (bottom). (d) Mean Fold Change Intensities of each individual method over single molecule RNA FISH signal intensities using the same fluorophore. Normalized from experiments taken to 12 rounds of amplification, including the experiment shown in Fig 1. Round 6 and round 12 converted intensities are normalized to clampFISH mean round 6 clampFISH intensity from this experiment to account for differences in exposure time (error bars show standard error of the mean over 2 biological replicates). ClampFISH leads to much higher signal intensity than the maximum signal intensity from the other methods. Hybridization chain reaction showed some background in the negative controls.
Supplementary Figure 13 Proximity dependence of clampFISH arms.
(a) Fluorescent signal from clampFISH probes targeting GFP mRNA on WM983bcells expressing GFP. Arms are hybridized adjacent to each other and clicked (bottom) Fluorescent signal remaining on the same cells after stripping with 60% formamide. (b) Fluorescent signal from clampFISH probes targeting GFP mRNA on WM983bcells expressing GFP. Arms are adjacent to each other without clicking (bottom) Fluorescent signal remaining on the same cells after stripping with 60% formamide. (c) Fluorescent signal from clampFISH probes targeting GFP mRNA on WM983bcells expressing GFP. One arm is specific to the GFP target while the other is non-targeting and the sample is clicked (bottom) Fluorescent signal remaining on the same cells after stripping with 60% formamide. (d) Fluorescent signal from clampFISH probes targeting GFP mRNA on WM983b cells that are not expressing GFP. Arms are hybridized adjacent to each other and clicked (bottom) Fluorescent signal remaining on the same cells after stripping with 60% formamide.
Supplementary Figure 14 DNA clampFISH optimization.
(a) DNA clampFISH probes were designed with different arm lengths: 15mer (same as RNA clampFISH), 23mer, and 30mer. The probes were tested on cells treated with and without RNase. (b) HIST1H4E mRNA clampFISH was performed to round 7 and the terminating backbone probed with a single-molecule FISH probe labeled in Alexa-488 (left). These cells were then pretreated, denatured and probed for 5S rDNA and the RNA signal assessed after treatment (right).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 (PDF 2774 kb)
Supplementary Protocol
ClampFISH working protocol (PDF 343 kb)
Supplementary Table
Supplementary Table 1 (XLSX 27 kb)
Rights and permissions
About this article
Cite this article
Rouhanifard, S., Mellis, I., Dunagin, M. et al. ClampFISH detects individual nucleic acid molecules using click chemistry–based amplification. Nat Biotechnol 37, 84–89 (2019). https://doi.org/10.1038/nbt.4286
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4286
This article is cited by
-
Decoder-seq enhances mRNA capture efficiency in spatial RNA sequencing
Nature Biotechnology (2024)
-
Highly sensitive spatial transcriptomics using FISHnCHIPs of multiple co-expressed genes
Nature Communications (2024)
-
Recent progress in co-detection of single-cell transcripts and proteins
Nano Research (2024)
-
The spatial organization of transcriptional control
Nature Reviews Genetics (2023)
-
HT-smFISH: a cost-effective and flexible workflow for high-throughput single-molecule RNA imaging
Nature Protocols (2023)