Abstract
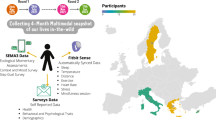
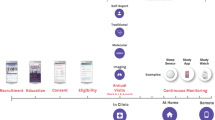
Personal data for 108 individuals were collected during a 9-month period, including whole genome sequences; clinical tests, metabolomes, proteomes, and microbiomes at three time points; and daily activity tracking. Using all of these data, we generated a correlation network that revealed communities of related analytes associated with physiology and disease. Connectivity within analyte communities enabled the identification of known and candidate biomarkers (e.g., gamma-glutamyltyrosine was densely interconnected with clinical analytes for cardiometabolic disease). We calculated polygenic scores from genome-wide association studies (GWAS) for 127 traits and diseases, and used these to discover molecular correlates of polygenic risk (e.g., genetic risk for inflammatory bowel disease was negatively correlated with plasma cystine). Finally, behavioral coaching informed by personal data helped participants to improve clinical biomarkers. Our results show that measurement of personal data clouds over time can improve our understanding of health and disease, including early transitions to disease states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hood, L. & Flores, M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N. Biotechnol. 29, 613–624 (2012).
Hood, L. & Friend, S.H. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat. Rev. Clin. Oncol. 8, 184–187 (2011).
Collins, F.S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Yong, P.L., Saunders, R.S. & Olsen, L. The Healthcare Imperative: Lowering Costs and Improving Outcomes: Workshop Series Summary (National Academies Press, 2010).
David, L.A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307 (2012).
Smarr, L. Quantifying your body: a how-to guide from a systems biology perspective. Biotechnol. J. 7, 980–991 (2012).
Hood, L. & Price, N.D. Promoting wellness and demystifying disease: The 100K project. Clinical OMICs 1, 20–23 (2014).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Girvan, M. & Newman, M.E.J. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 99, 7821–7826 (2002).
Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 103, 8577–8582 (2006).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Koh, K.K., Park, S.M. & Quon, M.J. Leptin and cardiovascular disease: response to therapeutic interventions. Circulation 117, 3238–3249 (2008).
Ridker, P.M. Clinical application of C-reactive protein for cardiovascular disease detection and prevention. Circulation 107, 363–369 (2003).
Woo, Y.C., Xu, A., Wang, Y. & Lam, K.S.L. Fibroblast growth factor 21 as an emerging metabolic regulator: clinical perspectives. Clin. Endocrinol. 78, 489–496 (2013).
Duong, T., Pommier, E.C. & Scheibel, A.B. Immunodetection of the amyloid P component in Alzheimer's disease. Acta Neuropathol. 78, 429–437 (1989).
Jenny, N.S., Arnold, A.M., Kuller, L.H., Tracy, R.P. & Psaty, B.M. Serum amyloid P and cardiovascular disease in older men and women: results from the Cardiovascular Health Study. Arterioscler. Thromb. Vasc. Biol. 27, 352–358 (2007).
Althaus, B.U., Staub, J.J., Ryff-De Lèche, A., Oberhänsli, A. & Stähelin, H.B. LDL/HDL-changes in subclinical hypothyroidism: possible risk factors for coronary heart disease. Clin. Endocrinol. 28, 157–163 (1988).
Jensen, L.J. et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 (2009).
Li, N., Wallén, N.H., Ladjevardi, M. & Hjemdahl, P. Effects of serotonin on platelet activation in whole blood. Blood Coagul. Fibrinolysis 8, 517–523 (1997).
Sauer, W.H., Berlin, J.A. & Kimmel, S.E. Selective serotonin reuptake inhibitors and myocardial infarction. Circulation 104, 1894–1898 (2001).
Li, M. et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 105, 2117–2122 (2008).
Poesen, R. et al. Microbiota-derived phenylacetylglutamine associates with overall mortality and cardiovascular disease in patients with CKD. J. Am. Soc. Nephrol. 27, 3479–3487 (2016).
Manichanh, C. et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211 (2006).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Bradley, R., Fitzpatrick, A.L., Jenny, N.S., Lee, D.-H. & Jacobs, D.R. Jr. Associations between total serum GGT activity and metabolic risk: MESA. Biomark. Med. 7, 709–721 (2013).
Lim, J.-S., Lee, D.-H., Park, J.-Y., Jin, S.-H. & Jacobs, D.R. Jr. A strong interaction between serum gamma-glutamyltransferase and obesity on the risk of prevalent type 2 diabetes: results from the Third National Health and Nutrition Examination Survey. Clin. Chem. 53, 1092–1098 (2007).
Lang, R., Gundlach, A.L. & Kofler, B. The galanin peptide family: receptor pharmacology, pleiotropic biological actions, and implications in health and disease. Pharmacol. Ther. 115, 177–207 (2007).
Prentice, K.J. et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 19, 653–666 (2014).
Zheng, J.S. et al. Serum metabolomics profiles in response to n-3 fatty acids in Chinese patients with type 2 diabetes: a double-blind randomised controlled trial. Sci. Rep. 6, 29522 (2016).
Guan, W. et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet 7, 321–331 (2014).
Kang, T.-W. et al. Genome-wide association of serum bilirubin levels in Korean population. Hum. Mol. Genet. 19, 3672–3678 (2010).
Rothman, N. et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 42, 978–984 (2010).
Okkels, H., Sigsgaard, T., Wolf, H. & Autrup, H. Arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) polymorphisms in susceptibility to bladder cancer: the influence of smoking. Cancer Epidemiol. Biomarkers Prev. 6, 225–231 (1997).
Diabetes Prevention Program Research Group. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. The Lancet 374, 1677–1686 (2009).
Adams, P.C. et al. Hemochromatosis and iron-overload screening in a racially diverse population. N. Engl. J. Med. 352, 1769–1778 (2005).
Gaich, G. et al. The effects of LY2405319, an FGF21 analog, in obese human subjects with type 2 diabetes. Cell Metab. 18, 333–340 (2013).
Meier, C. et al. TSH-controlled L-thyroxine therapy reduces cholesterol levels and clinical symptoms in subclinical hypothyroidism: a double blind, placebo-controlled trial (Basel Thyroid Study). J. Clin. Endocrinol. Metab. 86, 4860–4866 (2001).
Ruttmann, E. et al. Gamma-glutamyltransferase as a risk factor for cardiovascular disease mortality: an epidemiological investigation in a cohort of 163,944 Austrian adults. Circulation 112, 2130–2137 (2005).
Thompson, G.A. & Meister, A. Interrelationships between the binding sites for amino acids, dipeptides, and gamma-glutamyl donors in gamma-glutamyl transpeptidase. J. Biol. Chem. 252, 6792–6798 (1977).
Soga, T. et al. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J. Hepatol. 55, 896–905 (2011).
Rogers, A.J. et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One 9, e87538 (2014).
Sido, B. et al. Impairment of intestinal glutathione synthesis in patients with inflammatory bowel disease. Gut 42, 485–492 (1998).
Guo, L. et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc. Natl. Acad. Sci. USA 112, E4901–E4910 (2015).
Hood, L. & Price, N.D. Demystifying disease, democratizing health care. Sci. Transl. Med. 6, 225ed5 (2014).
Tangpricha, V., Pearce, E.N., Chen, T.C. & Holick, M.F. Vitamin D insufficiency among free-living healthy young adults. Am. J. Med. 112, 659–662 (2002).
Micheel, C.M., Nass, S.J. & Omenn, G.S. Evolution of Translational Omics: Lessons Learned and the Path Forward (National Academies Press, 2012).
Glusman, G. et al. Identification of copy number variants in whole-genome data using Reference Coverage Profiles. Front. Genet. 6, 45 (2015).
Glusman, G., Caballero, J., Mauldin, D.E., Hood, L. & Roach, J.C. Kaviar: an accessible system for testing SNV novelty. Bioinformatics 27, 3216–3217 (2011).
Caporaso, J.G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Rideout, J.R. et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ 2, e545 (2014).
Whittaker, R.H. Evolution and measurement of species diversity. Taxon 21, 213–251 (1972).
Caporaso, J.G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
McDonald, D. et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Lane, D.J. in Nucleic Acid Techniques in Bacterial Systematics (eds. Stackebrandt, E. & Goodfellow, M.) 115–175 (John Wiley and Sons, Chichester, UK, 1991).
Lozupone, C., Lladser, M.E., Knights, D., Stombaugh, J. & Knight, R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 (2011).
Hamady, M., Lozupone, C. & Knight, R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2010).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Pérez, F. & Granger, B.E. IPython: a system for interactive scientific computing. Comput. Sci. Eng. 9, 21–29 (2007).
Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Kusebauch, U. et al. Human SRMAtlas: A resource of targeted assays to quantify the complete human proteome. Cell 166, 766–778 (2016).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with python. Proceedings of the 9th Python in Science Conference (eds. van der Walt, S. & Millman, J.) 57–61 (SciPy, 2010).
Schult, D.A. & Swart, P. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science Conference (eds. Varoquaux, G., Vaught, T. & Millman, J.) 11–15 (SciPy, 2008).
Green, R.C. et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15, 565–574 (2013).
Acknowledgements
We would like to acknowledge significant contributions to this study from our 108 Pioneers, S. Kaplan, S. Mecca, S. Bell, G. Sorensen, C. Lewis, T. Kilgallon, M. Brunkow, S. Huang, C.-Y. Huang, D. Mauldin, S. Speck, M. Raff, J. Pizzorno, J. Guiltinan, R. Green, L. Smarr, E. Lazowska, C. Witwer, M. Flores, and many others who helped us on this wellness journey. This work was supported in part by the Robert Wood Johnson Foundation (L.H., N.D.P.), the M.J. Murdock Charitable Trust (L.H., N.D.P.), NIH grants 2P50GM076547 (L.H., N.D.P.), P30ES017885 (G.S.O.), U24CA2210967 (G.S.O.), RC2HG005805 (R.L.M.), and Arivale.
Author information
Authors and Affiliations
Contributions
L.H. and N.D.P. conceived of and led the study. J.C.L. designed and managed the clinical and coaching aspects of the study. A.T.M. and J.C.E. performed most of the computational analyses. G.G. contributed many important ideas from the beginning of the study. G.G., R.L., and D.T.M. performed additional computational analysis. N.D.P., A.T.M., J.C.E., G.G., R.L., D.T.M., G.S.O., J.C.L., and L.H. analyzed data. C.L. generated the Olink proteomics data. U.K., C.L.M., Y.Z., S.Q., and R.L.M. generated the mass spectrometry proteomics data. K.B. managed most of the logistics of implementing the study. A.T.M., N.D.P., and L.H. were the primary writers of the paper, with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
L.H. and N.D.P. are co-founders of Arivale and hold stock in the company. N.D.P. is on the Arivale board of directors; L.H. is chair and G.S.O. a member of Arivale's scientific advisory board. A.T.M., J.C.E., K.B., and J.C.L. are employees of Arivale and have stock options in the company, as do G.G. and G.S.O.
Integrated supplementary information
Supplementary Figure 1 Full inter-omic cross-sectional correlation network
Statistically-significant inter-omic cross-sectional Spearman correlations (padj<0.05) between all datasets collected in our cohort.
Supplementary Figure 2 Modularity vs. community analysis iteration
The maximum modularity observed in our inter-omic cross-sectional community analysis was 0.386 at iteration 61 of community pruning. There were 267 total iterations of community analysis.
Supplementary Figure 3 Recruitment, onboarding, and other important events in the P100
Flowchart of important events in the P100, including recruitment, onboarding, withdrawals, data collection, coaching calls, and events.
Supplementary Figure 4 Genetic risk factors for hemochromatosis and ferritin levels
Boxplots for ferritin levels of male (A) and female (B) participants by round in the P100. Only one male in our study was homozygous for 282YY and was diagnosed with hemochromatosis after physician referral. Changes in ferritin levels are shown by the red arrows. A second male who was heterozygous for both risk factors (282YC/63DH) did not receive therapeutic phlebotomy, and ferritin levels increased. Six other males presented at baseline (round 1) with elevated ferritin levels but neither of these genetic risk factors; they were referred to their physician for monitoring. Four of the six were of self-reported Asian ancestry (orange dots).
Supplementary Figure 5 Population distribution of the 108 Pioneers (PC2 vs PC3)
PCA using a sample of 250,000 common SNPs. Translucent colored points represent the 2504 individuals in the Thousand Genomes Project, phase 3, color-coded by population. Black points represent the 108 Pioneers. AFR, EUR, SAS, EAS, and AMR represent the continental populations: African, European, South Asian, East Asian and Admixed Americans, respectively.
Supplementary Figure 6 Population distribution of the 108 Pioneers (PC1 vs PC2)
PCA using a sample of 250,000 common SNPs. Translucent colored points represent the 2504 individuals in the Thousand Genomes Project, phase 3, color-coded by population. Black points represent the 108 Pioneers. AFR, EUR, SAS, EAS, and AMR represent the continental populations: African, European, South Asian, East Asian and Admixed Americans, respectively.
Supplementary Figure 7 Dose-dependent effects of vitamin D supplementation
A common intervention for our participants was vitamin D supplementation. The Institute of Medicine has recommended a minimum 25-hydroxyvitamin D level of 20 ng/mL, while the Endocrine Society recommends a minimum level of 30 ng/mL. Shown here are the dose-dependent effects of supplementation on 25-hydroxyvitamin D levels from round 1 to round 2, with individuals taking less than 3000 IUs/day exhibiting relatively little gains in 25-hydroxyvitamin D levels. Individuals that were noncompliant with the recommendations (N=13) made no gains in 25-hydroxyvitamin D levels. A one-way ANOVA yields p=0.004 that a significant difference exists between one of the groups. Significant differences with noncompliant participants at the p<0.05 level are indicated with asterisks, as determined by Tukey’s range test. Individuals with low blood levels of 25-hydroxyvitamin D were recommended doses between 1000 and 5000 IU. If over time blood levels did not increase at the highest doses, individuals were referred to their physician for evaluation and, if appropriate, higher doses.
Supplementary Figure 8 Gut microbiome stability over nine months
Participant microbiomes tend to resemble themselves over time. Plotted in red is the unweighted UniFrac distance between consecutive microbiome samples for all participants. The blue box-and-whisker plots represent the distance distribution between each sample and all others in the same time points. In 97% of cases, an individual’s cross-timepoint distance is lower than the median inter-individual distance.
Supplementary Figure 9 Correlation across different vendors
Most clinical laboratory measurements were assayed by only one of the lab vendors (Quest or Genova) but certain measurements were measured by both due to overlaps in the standard analysis panels. Additionally, some analytes from the metabolomics and proteomics data were also measured by the clinical labs. This figure shows a comparison of these duplicated analytes. For example, triglycerides, total cholesterol, and fasting glucose show high correlation between clinical lab vendors, while LDL particle number is less correlated. The correlations represented in this figure have been removed from our correlation networks.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 6–7, 9–13 (PDF 2297 kb)
Supplementary Code
Supplementary Code zip (ZIP 72 kb)
Supplementary Table 1
All analytes measured in the P100 (XLSX 317 kb)
Supplementary Table 2
Complete inter-omic correlation network for cross-sectional correlations (XLSX 247 kb)
Supplementary Table 3
Complete intra- and inter-omic correlation network for cross-sectional correlations (XLSX 1240 kb)
Supplementary Table 4
Complete inter-omic correlation network for delta correlations (XLSX 191 kb)
Supplementary Table 5
Complete intra- and inter-omic correlation network for delta correlations (XLSX 1834 kb)
Supplementary Table 8
Polygenic score quantitative traits tested in the P100 (XLSX 51 kb)
Supplementary Table 14
Age and sex adjustments for the correlation networks (XLSX 81 kb)
Rights and permissions
About this article
Cite this article
Price, N., Magis, A., Earls, J. et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol 35, 747–756 (2017). https://doi.org/10.1038/nbt.3870
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3870
This article is cited by
-
The transition from genomics to phenomics in personalized population health
Nature Reviews Genetics (2024)
-
A Proactive Intervention Study in Metabolic Syndrome High-Risk Populations Using Phenome-Based Actionable P4 Medicine Strategy
Phenomics (2024)
-
Reference Materials for Improving Reliability of Multiomics Profiling
Phenomics (2024)
-
Predicting disease severity in multiple sclerosis using multimodal data and machine learning
Journal of Neurology (2024)
-
Protocol of a parallel, randomized controlled trial on the effects of a novel personalized nutrition approach by artificial intelligence in real world scenario
BMC Public Health (2023)