Abstract
The potential of the diverse chemistries present in natural products (NP) for biotechnology and medicine remains untapped because NP databases are not searchable with raw data and the NP community has no way to share data other than in published papers. Although mass spectrometry (MS) techniques are well-suited to high-throughput characterization of NP, there is a pressing need for an infrastructure to enable sharing and curation of data. We present Global Natural Products Social Molecular Networking (GNPS; http://gnps.ucsd.edu), an open-access knowledge base for community-wide organization and sharing of raw, processed or identified tandem mass (MS/MS) spectrometry data. In GNPS, crowdsourced curation of freely available community-wide reference MS libraries will underpin improved annotations. Data-driven social-networking should facilitate identification of spectra and foster collaborations. We also introduce the concept of 'living data' through continuous reanalysis of deposited data.
Similar content being viewed by others
Main
NP from marine and terrestrial environments, including their inhabiting microorganisms, plants, animals, and humans, are routinely analyzed using MS. However, a single MS experiment can collect thousands of MS/MS spectra in minutes1, and individual projects can acquire millions of spectra. These data sets are too large for manual analysis. Furthermore, comprehensive software and proper computational infrastructure are not readily available and only low-throughput sharing of either raw or annotated spectra is feasible, even among members of the same laboratory. The potentially useful information in MS/MS data sets can thus remain buried in papers, laboratory notebooks, and private databases, hindering retrieval, mining, and sharing of data and knowledge. Although several NP databases—Dictionary of Natural Products2, AntiBase3, and MarinLit4—assist in dereplication (identification of known compounds), these resources are not freely available and do not process MS data. Conversely, MS databases, including MassBank5, Metlin6, mzCloud7, and ReSpect8, host MS/MS spectra but limit data analyses to several individual spectra or a limited amount of liquid chromatography (LC)–MS files. Other free online computation resources that leverage the MS/MS spectra of Metlin, such as those provided by mzCloud and XCMS Online, are available. However, neither of those allows free download of its reference library.
Global genomics and proteomics research has been facilitated by the development of integral resources, such as the US National Center for Biotechnology Information (NCBI; Bethesda, MD, USA) and UniProt KnowledgeBase (UniProtKB), which provide robust platforms for data sharing and knowledge dissemination9,10. Recognizing the need for an analogous community platform to analyze NP MS data, we present GNPS. GNPS is a data-driven platform for the storage, analysis, and knowledge dissemination of MS/MS spectra that enables community sharing of raw spectra, continuous annotation of deposited data, and collaborative curation of reference spectra (referred to as spectral libraries) and experimental data (organized as data sets).
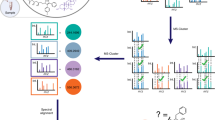
GNPS provides the ability to analyze a data set and to compare it to all publicly available data. By building on the computational infrastructure of the University of California San Diego (UCSD) Center for Computational Mass Spectrometry (CCMS; http://proteomics.ucsd.edu/), GNPS provides public data set deposition and/or retrieval through the Mass Spectrometry Interactive Virtual Environment (MassIVE) data repository. The GNPS analysis infrastructure further enables online dereplication6,11,12,13, automated molecular networking analysis14,15,16,17,18,19,20,21, and crowdsourced MS/MS spectrum curation. Each data set added to the GNPS repository is automatically reanalyzed in the next monthly cycle of continuous identification (see 'Living data by continuous analysis' below). Each of these tens of millions of spectra in GNPS data sets is matched to reference spectral libraries to annotate molecules and to discover putative analogs (Fig. 1a). From January 2014 to November 2015, GNPS grew to serve 9,267 users from 100 countries (Fig. 1b), with 42,486 analysis sessions that have processed >93 million spectra as molecular networks from a quarter-million LC–MS runs. Searches against a combined catalog of over 221,000 MS/MS reference library spectra from 18,163 compounds (Supplementary Table 1) are possible, and GNPS has matched almost one hundred million MS/MS spectra in all public and private search jobs using an estimated 84,000 compute hours.
(a) Representation of interactions among the NP community, GNPS spectral libraries, and GNPS data sets. At present 221,083 MS/MS spectra from 18,163 unique compounds are used for searches in GNPS. These include both third-party libraries, such as MassBank, ReSpect, and NIST, as well as spectral libraries created for GNPS (GNPS-Collections) and spectra from the NP community (GNPS-Community). GNPS spectral libraries grow through user contributions of new identifications of MS/MS spectra. To date, 55 community members have contributed 8,853 MS/MS spectra from 5,568 unique compounds (30.5% of the unique compounds available). In addition, ongoing curation efforts have already yielded 563 annotation updates for library spectra. The utility of these libraries is to dereplicate compounds (recognition of previously characterized and studied known compounds), in both public and private data. This dereplication process is performed on all public data sets and results are automatically reported, thus enabling users to query all data sets, organisms, and conditions. Automatic reanalysis of all public data creates a virtuous cycle in which contributions to libraries can be matched to all public data. Combined with molecular networking (Fig. 3), this automatic reanalysis empowers community members to identify analogs that can then be added to GNPS spectral libraries. (b) The GNPS platform has grown to serve a global user base of >9,200 users from 100 countries.
GNPS spectral libraries
GNPS spectral libraries enable dereplication, variable dereplication (approximate matches to spectra of related molecules), and identification of spectra in molecular networks. GNPS has collected available MS/MS spectral libraries relevant to NP (which also include other metabolites and molecules), including MassBank5, ReSpect8, and NIST22 (Table 1, Fig. 2a and Supplementary Table 1). Altogether, these third-party libraries total 212,230 MS/MS spectra representing 12,694 unique compounds (Fig. 2b). Although this combined collection of reference spectra provides a starting point for dereplication, only 1.01% of all spectra in public GNPS data sets has been matched to this collection, indicating insufficient chemical space coverage. Although the NP community is working to populate this 'missing' chemical space, there is no way to report discoveries of chemistries in an easily verifiable and reusable format.
(a) The computational resources of the metabolomics and the NP community fall into two main categories: first, reference collections (red dots) of MS/MS spectral libraries; and second, data repositories (blue dots) designed to publicly share raw MS data associated with research projects. Reference collection resources are contributors and aggregators of reference MS/MS spectra, some of which also include data analysis tools, for example, online multi-spectrum MS/MS search (magnifying glass icon). Several resources have aggregated MS/MS spectra from various reference collections so that the analysis tools at a respective resource can leverage more of the community efforts to annotate data (red and blue arrows). GNPS has imported all freely available reference collections (>221,000 MS/MS spectra) and makes them available for online analyses. GNPS and several other resources provide both reference MS/MS spectra and data in an open and free manner to the public (pink caps). (b) Comparison of spectral library sizes of available libraries (MassBank, ReSpect, and NIST) and GNPS libraries; GNPS-Collections includes newly acquired spectra from synthetic or purified compounds and GNPS-Community includes all community-contributed spectra. (c) Searching all public GNPS data sets revealed that MassBank, ReSpect, and NIST libraries matched to 1,217 unique compounds, with GNPS libraries increasing unique compound matches by 41% (corresponding to 29% of total unique matches) with an accompanying 4% increase in spectral library size. Overall, GNPS libraries increase the total number of spectra matched in public data sets by 144% (59% of total public MS/MS matches), and spectra matches across all GNPS public and private data by 767% (88% of all MS/MS matches). (d) The distribution of precursor masses in all GNPS public data sets is shown in gray and compared to the precursor mass distributions of MassBank, ReSpect, NIST, and GNPS libraries (color key as in b). Though GNPS libraries have a combined size that is smaller than MassBank, ReSpect, and NIST, GNPS libraries have a higher proportion of molecules in the higher m/z range and therefore complement the proportionately lower precursor mass molecules in other libraries. (e) The quality of spectrum matches obtained by searching against the available spectral libraries is assessed by user ratings (1 to 4 stars; Supplementary Table 6) of continuous identification results. User ratings of >2.5 stars for >98% of GNPS library matches compares favorably with the 90% mark for NIST matches, whose high marks demonstrate how important these third-party libraries still are to the GNPS platform. We note that the lower mark for NIST matches does not suggest lower-quality spectra. It is more likely explained by its higher emphasis on lower precursor mass molecules with spectra that have fewer peaks and are generally harder to match.
To begin to address this pressing need, GNPS houses both newly acquired reference spectra (GNPS-Collections) as well as a crowdsourced library of community-contributed reference spectra (GNPS-Community). The GNPS-Collections data set includes NP and pharmacologically active compounds, totaling 6,629 MS/MS spectra of 4,243 compounds (Fig. 2b, Supplementary Table 1, Supplementary Notes 1 and 2, and Supplementary Table 2). The GNPS-Community library has grown to include 2,224 MS/MS spectra of 1,325 compounds from 55 worldwide contributors. Although the total number of MS/MS spectra in GNPS libraries is only 4% of the MS/MS spectra collected in third-party libraries, GNPS libraries contribute matches of MS/MS spectra at a scale disproportionate to their size (Fig. 2c). The GNPS libraries account for 29% of unique compound matches and 59% of the MS/MS matches in public (88% of public and private) data. This indicates that the GNPS libraries contain compounds that are complementary to the chemical space represented in other libraries (Fig. 2c,d). Moreover, in contrast to third-party libraries, spectra submitted to GNPS-Community libraries are immediately searchable by the whole community, such that submissions seamlessly transfer knowledge between laboratories (Fig. 1a) in a process that is akin to the addition of genome annotations to GenBank9.
To create a robust library, we have to ensure that submissions are peer-reviewed and, if necessary, annotations corrected or updated as appropriate. Reference spectra submitted to the GNPS-Community library are categorized by the estimated reliability of the proposed submissions. Gold reference spectra must be derived from structurally characterized synthetic or purified compounds and can be submitted only by approved users. Approval is given to contributors who have undergone training. Training is initiated by contacting the corresponding authors or CCMS administrators. Silver reference spectra need to be supported by an associated publication, and bronze reference spectra comprise all remaining putative annotations (Supplementary Table 3). This type of division of spectra is reminiscent of RefSeq/TPA/GenBank9,23 (genomics) and Swiss-Prot/TrEMBL/UniProt24,25 (proteomics), allowing varying tradeoffs between comprehensiveness and reliability of annotations defined as gold, silver, or bronze (Fig. 2e).
To enable refinements or corrections of annotations, GNPS allows community-driven, iterative re-annotation of reference MS/MS spectra in a wiki-like fashion, to progressively improve the library and converge toward consensus annotation of all MS/MS spectra of interest. This is a process similar to the iterative annotation of the human genome9. To date, 563 annotation revisions have been made in GNPS (Supplementary Table 4), most of which added metadata to library spectra or refined compound names. The history of each annotation is retained so that users can discuss the proper annotation and address disagreements through comment threads.
Dereplication using GNPS
High-throughput dereplication of NP MS/MS data is implemented in GNPS by querying newly acquired MS/MS spectra against all the accumulated reference spectra in GNPS spectral libraries (Fig. 3a). To date, >93 million MS/MS spectra from various instruments (including Orbitrap, Ion Trap, qTOF, and FT-ICR) have been searched at GNPS, yielding putative dereplication matches of 7.7 million spectra to 15,477 compounds. In the second stage of dereplication, GNPS goes beyond re-identification by using variable dereplication, which is a modification-tolerant spectral library search that is mediated by a spectral alignment algorithm. Variable dereplication enables the detection of significant matches to either putative analogs of known compounds (e.g., differing by one modification or substitution of a chemical group) or compounds belonging to the same general class of molecules (Fig. 3b). Variable dereplication is not available through any other computational platform. For example, GNPS variable dereplication has detected compounds with different levels of glycosylation on various substrates. As MS/MS fragmentation preferentially results in peaks from glycan fragments, it is possible to detect sets of compounds with related glycans even when the substrates to which the glycans are attached are themselves unrelated26. To date, 3,891 putative analogs have been identified in public data using GNPS variable dereplication (Supplementary Table 5). These 3,891 putative analogs include several unique molecules that could be user-curated and added to GNPS reference libraries (see 'Molecular Explorer' below on accessing and annotating putative analogs).
(a) Molecular networks are constructed from the alignment of MS/MS spectra to one another. Edges connecting nodes (MS/MS spectra) are defined by a modified cosine scoring scheme that determines the similarity of two MS/MS spectra with scores ranging from 0 (totally dissimilar) to 1 (completely identical). MS/MS spectra are also searched against GNPS spectral libraries, seeding putative node matches in the molecular networks. Networks are visualized online in-browser or exported for third-party visualization software such as Cytoscape31. (b) An example alignment between three MS/MS spectra of compounds with structural modifications that are captured by modification-tolerant spectral matching used in variable dereplication and molecular networking. (c) In-browser molecular network visualization enables users to interactively explore molecular networks without requiring any external software. To date, >11,000 molecular networks have been analyzed using this feature. Within this interface, (i) users are able to define cohorts of input data and correspondingly, nodes within the network are represented as pie charts to visualize spectral count differences for each molecule across cohorts. (ii) Node labels indicate matches made to GNPS spectral libraries, with additional information displayed with mouseovers. These matches provide users a starting point to annotate unidentified MS/MS spectra within the network. (iii) To facilitate identification of unknowns, users can display MS/MS spectra in the right panels by clicking on the nodes in the network, giving direct interactive access to the underlying MS/MS peak data. Furthermore, alignments between spectra are visualized between spectra in the top right and bottom right panels to gain insight as to what underlying characteristics of the molecule could elicit fragmentation perturbations.
To assess the reliability of the MS/MS matches found by GNPS dereplication, GNPS users can rate the quality of matches returned by automated GNPS reanalysis (see below). These ratings are four star (correct), three star (likely correct; e.g., could also be isomers with similar fragmentation patterns), two star (unable to confirm the annotation due to limited information), and one star (incorrect) (Supplementary Table 6). So far, of the 3,608 matches that have been rated, 139 (3.9%) matches were given one or two stars (insufficient information (2.9%) or incorrect (1%)) by user ratings. These percentages are consistent with the false-discovery rates estimated using spectral library searches of benchmark LC–MS data sets with compound standards (Supplementary Note 3, Supplementary Figs. 1 and 2, and Supplementary Table 7). Furthermore, these 3,608 match ratings were associated with 2,041 library spectra, therefore, the average rating of a library spectrum can offer insight into the reliability of its reference annotation, not unlike Yelp ratings for restaurants. Incorrect matches can arise through either spurious high-scoring matches to library spectra or incorrect annotations for library spectra. Of the 2,041 library spectra with match ratings, 72 (3.5%) of spectra had average ratings below 2.5 stars. These percentage ratings were further broken down by spectral library (Fig. 2e). We found that for GNPS-Collection and GNPS-Community libraries, only 29 out of 1,746 (1.7%) of the rated library spectra had average ratings below 2.5 stars. These ratings demonstrate that the perceived reliability of GNPS spectral libraries compares favorably with established community resources such as NIST and MassBank, in which 10.5% and 20.1% of the ratings were below 2.5 stars, respectively, and provides confidence that the community curation process is robust and that third-party libraries integrate well with GNPS. The main advantages of searching using GNPS are the option to run simple or variable dereplication against all publicly accessible reference spectra, and that community-rated matches can be used to improve the quality of the reference libraries and matching algorithms. These dereplication capabilities are not possible with existing published resources.
Molecular networking
Molecular networks are visual displays of the chemical space present in MS experiments. GNPS can be used for molecular networking14,15,16,17,18,19,20,21,27,28, a spectral correlation and visualization approach that can detect sets of spectra from related molecules (so-called spectral networks29), even when the spectra themselves are not matched to any known compounds (Fig. 3a). Spectral alignment15,27 detects similar spectra from structurally related molecules, assuming these molecules fragment in similar ways reflected in their MS/MS patterns (Fig. 3b), analogous to the detection of related protein or nucleotide sequences by sequence alignment.
GNPS is currently the only public infrastructure that enables molecular networking. The visualization of molecular networks in GNPS represents each spectrum as a node, and spectrum-to-spectrum alignments as edges (connections) between nodes. Nodes can be supplemented with metadata, including dereplication matches or information that is provided by the user, such as abundance, origin of product, biochemical activity or hydrophobicity, which can be reflected in a node's size or color. It is possible to visualize the map of related molecules as a molecular network21,30,31,32,33 (Supplementary Fig. 3) online at GNPS (Fig. 3c) or exported for analysis in Cytoscape31. Molecular networking analyses of 272 public data sets (Fig. 4a) from a diverse range of samples reveal that on average 35.2% of all unidentified nodes are matched to other spectra of related molecules within a cosine score of 0.8 (44.7% of all nodes in more exploratory networks with a cosine score of 0.65; Supplementary Table 8). This suggests that a large fraction of all unidentified spectra would be identifiable if their or their neighboring nodes' reference spectra were available in the reference spectral libraries.
(a) A global snapshot of the state of MS/MS matching of public NP data sets available in GNPS using molecular networking and library search tools. Identified molecules (1.9% of the data) are MS/MS spectrum matches to library spectra with a cosine >0.7. Putative analog molecules (another 1.9% of the data) are MS/MS spectra that are not identified by library search but rather are immediate neighbors of identified MS/MS spectra in molecular networks. Identified Networks (9.9% of the data) are connected components within a molecular network that have at least one spectrum match to library spectra. Unidentified networks (25.2% of the data) are molecular networks where none of the spectra match to library spectra; these networks potentially represent compound classes that have not yet been characterized. Exploratory networks (an additional 20.1% of the data) are unidentified connected components in molecular networks with more relaxed parameters (Supplementary Table 8). Thus, 55.3% of the MS/MS spectra at least have one related MS/MS spectrum in spectral networks, with 44.7% having none. In this 44.7% of the data, each MS/MS spectrum has been observed in two separate instances and should not constitute noise. Altogether, this analysis indicates that most of the chemical space captured by MS remains unexplored. (b) In the past year, there has been substantial growth in the GNPS spectral libraries, driving an increase in the match rates of all public data. The number of unique compounds matched in the public data has increased tenfold; the number of total spectra matched has increased 22-fold; and the average match rate has increased threefold. It is expected that identification rates will continue to grow with further contributions from the community to the GNPS-Community spectral library.
Living data by continuous analysis
Funding agencies and publishers have called for raw scientific data, including MS data, and analysis methods to be made publicly available where possible. Consistent with this aim, GNPS data sets usually comprise the full set of MS files produced during a NP research project or the full set of spectra analyzed for a peer-reviewed publication (Supplementary Note 4). Although it is potentially advantageous to the community for all data to be made public, GNPS user data can remain private until users explicitly choose to make them public (private data are also analyzable and privately sharable, with >93 million spectra in >250,000 private LC–MS runs already searched using GNPS). GNPS has the largest collection of publicly accessible natural product and metabolomics MS/MS data sets and is the only infrastructure where public data sets can be reanalyzed together and compared with each other (Table 1). To date, GNPS has made 272 public GNPS data sets openly available, which comprise >30,000 MS runs with ∼84 million MS/MS spectra. In common with other public repositories34,35, GNPS data sets can be downloaded. However, data availability on its own does not suffice to enable data reuse. GNPS is unique among MS repositories by enabling continuous identification: the periodic and automated reanalysis of all public data sets (Supplementary Notes 5 and 6, and Supplementary Tables 9 and 10). This continuous reanalysis, which incorporates molecular networking and dereplication tools, implements a 'virtuous cycle' (Fig. 1a). Because GNPS spectral libraries are constantly growing, owing to community contributions and continued generation of reference spectra, the number of matches made by successive reanalyses of public data sets has already grown and is expected to continue to grow over time (Fig. 4b). GNPS users are periodically updated with alerts of new search results.
For example, a Streptomyces roseosporus project (MSV000078577) was deposited April 8, 2014. At first, only seven MS/MS spectra were matched. However, as of July 14, 2015, 36 spectral matches were made to GNPS libraries. Overall, the total number of compounds matched to GNPS data sets increased more than tenfold, whereas the number of matched MS/MS spectra in GNPS data sets increased >20-fold in 2015 (Fig. 4b). GNPS users can also subscribe to specific data sets of interest, rather like 'following' people on Twitter. When new matches are made, changed, or revoked, all subscribers are notified of new information by an e-mail summarizing changes in identification. From April 2014 to July 2015, 45 updates were initiated by CCMS and automatically sent to subscribers (Supplementary Fig. 4). Update e-mails have led to substantially more views per data set, compared with non-GNPS data sets (192 proteomics data sets deposited in MassIVE). Continuous identification not only keeps a single data set 'alive', it can also create connections between data sets and users over time. Similarities between data sets could form the basis of a data-mediated social network of users with potentially related research interests despite seemingly disparate research fields, rather like the 'People You May Know' feature on LinkedIn. On average, each GNPS user already has five suggested collaborators (Supplementary Fig. 5).
Molecular explorer
Molecular Explorer is a feature that can only be implemented on 'living data' repositories and thus exists only in GNPS. Molecular Explorer allows users to find all data sets and putative analogs that have ever been observed for a given molecule of interest. We anticipate that this feature could guide the discovery of previously unknown analogs of existing antibiotics. Public NP data contain >100 unidentified putative analogs of antibiotics, such as valinomycin, actinomycin, etamycin, hormaomycin, stendomycin, daptomycin, erythromycin, napsamycin, clindamycin, arylomycin, and rifamycin, highlighting a clear potential to generate leads to discover structurally related antibiotics through the application of GNPS (Supplementary Fig. 6, Supplementary Table 5 and Supplementary Note 7). Box 1 illustrates how this approach was applied to stenothricin (Fig. 5).
(a) The stenothricin molecular family was identified during analysis of a molecular network between chemical extracts of S. roseosporus NRRL 15998 (green) and Streptomyces sp. DSM5940 (blue). This analysis indicates that Streptomyces sp. DSM5940 produces a structurally similar compound to stenothricin with a −41 Da m/z difference. An enlarged version of the network can be found in Supplementary Figure 8. (b) Based on preliminary structural analysis, stenothricin-GNPS (41 Da) may contain a Lys to Ser substitution. (c) Comparison of the MS/MS of stenothricin D with stenothricin-GNPS 2. (d) Although structurally related, stenothricin and stenothricin-GNPS have different effects on E. coli as visualized using fluorescence microscopy. Red is the membrane stain FM4-64, blue is the membrane-permeable DNA stain DAPI (4′,6-diamidino-2-phenylindole), green is the membrane impermeable DNA stain SYTOX green. SYTOX green stains DNA only when the cell membrane is damaged. Scale bar, 2 μm.
Several published applications of molecular networking and MS/MS-based dereplication using GNPS have been reported while the infrastructure has been under development. Specifically, GNPS has enabled the discovery of NP including colibactin41,42,43,44,45, characterization of biosynthetic pathways46,47, understanding of the chemistry of ecological interactions28,48,49,50,51,52, and development of metabolomics bioinformatics methods53. The application of GNPS workflows to such diverse research areas demonstrates its utility.
Conclusions
GNPS provides a community-led knowledge space in which NP data can be shared, analyzed, and annotated by researchers worldwide. It enables a cycle of annotation in which users curate data, continuous dereplication enables product identification, and a knowledge base of reference spectral libraries and public data sets is created. Selected views from community members were sought by Nature Biotechnology and are presented, together with author responses, in Supplementary Note 8.
The transformation of deposited spectra into living data that are enabled by the GNPS platform could mediate connections between researchers and has the potential to transform data networks into social networks. Of 1,272 compound identifications obtained by continuous identification with the GNPS-Community library, 1,063 (83.6%) were made using reference spectra that were not uploaded by the submitter. In other words, the vast majority of identifications were enabled by other community members. This reuse of knowledge and data is analogous to other community-wide curation efforts including Wikipedia and crowdsourced dictionaries. From the time of their initial deposition, 59% of data sets have an increased number of identifications, with the average data set more than doubling the number of identifications since submission (Supplementary Fig. 19). GNPS enables facile sharing of individual analyses (Supplementary Fig. 20) and uses molecular networks to reveal connections among data sets from different laboratories and biological sources that would otherwise remain disconnected. To date, 3,145 analysis jobs have included files shared among GNPS users, encompassing 548 unique pairs of individuals' collaborations. GNPS recasts public data sets as 'conversation starters' in a data-mediated social network.
Although we have described only one simple application of GNPS in this Perspective (the identification of a stenothricin analog in Box 1), the community has already begun to use GNPS to expedite NP analysis28,41,43,45,46,50,52. Furthermore, we expect the user base of GNPS to expand to include other communities that use MS/MS data, including those studying metabolomes, microbiomes, exposomes (measurements of life-course environmental exposures), and the chemistry of the human habitat, or researchers involved in areas as diverse as drug discovery, biomarker stratification of patients and adsorption, distribution, metabolism, excretion and toxicology studies, food science, agricultural sciences, and ocean science, to name a few, all resulting in different GNPS workflows42,44,47,51,53.
Genomics9 and protein structure analysis54 have already shown that models of global collaboration and social cooperation can empower scientific communities to collectively translate big data into shared, reusable knowledge. We believe that GNPS will transform NP research in a similar manner, profoundly influencing the way we explore molecules using MS.
Additional details about the methods used in this work can be found in the Supplementary Methods. Source code and license are available at the CCMS software tools webpage as well as at GitHub (https://github.com/CCMS-UCSD). Source code is also available with this manuscript as Supplementary Source Code.
References
Bouslimani, A., Sanchez, L.M., Garg, N. & Dorrestein, P.C. Mass spectrometry of natural products: current, emerging and future technologies. Nat. Prod. Rep. 31, 718–729 (2014).
Dictionary of Natural Products http://dnp.chemnetbase.com/ (2013).
Laatsch, H. AntiBase 2012: The Natural Compound Identifiers (Wiley-VCH, 2011).
Blunt, J. & Munro, M. MarinLit: a database of the marine natural products literature http://pubs.rsc.org/marinlit/ (Department Chem. Univ. Canterbury, Canterbury, New Zealand) (2003).
Hisayuki, H. et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 (2010).
Smith, C.A. et al. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 (2005).
mzCloud: advanced mass spectral database https://www.mzcloud.org/.
Sawada, Y. et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry 82, 38–44 (2012).
Benson, D.A. et al. GenBank. Nucleic Acids Res. 41, D36–D42 (2013).
Magrane, M. & UniProt Consortium. UniProt Knowledgebase: a hub of integrated protein data. Database 2011, bar009 (2011).
Lang, G. et al. Evolving trends in the dereplication of natural product extracts: new methodology for rapid, small-scale investigation of natural product extracts. J. Nat. Prod. 71, 1595–1599 (2008).
Ito, T. & Masubuchi, M. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. (Tokyo) 67, 353–360 (2014).
Little, J.L., Williams, A.J., Pshenichnov, A. & Tkachenko, V. Identification of “known unknowns” utilizing accurate mass data and ChemSpider. J. Am. Soc. Mass Spectrom. 23, 179–185 (2012).
Moree, W.J. et al. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 109, 13811–13816 (2012).
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 109, E1743–E1752 (2012).
Nguyen, D.D. et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. USA 110, E2611–E2620 (2013).
Sidebottom, A.M., Johnson, A.R., Karty, J.A., Trader, D.J. & Carlson, E.E. Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. A.C.S. Chem. Biol. 8, 2009–2016 (2013).
Vizcaino, M.I., Engel, P., Trautman, E. & Crawford, J.M. Comparative metabolomics and structural characterizations illuminate colibactin pathway-dependent small molecules. J. Am. Chem. Soc. 136, 9244–9247 (2014).
Wilson, M.C. et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 506, 58–62 (2014).
Engel, P., Vizcaino, M.I. & Crawford, J.M. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl. Environ. Microbiol. 81, 1502–1512 (2015).
Yang, J.Y. et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 76, 1686–1699 (2013).
The National Institute of Standards and Technology. NIST standard reference database 1A http://www.nist.gov/srd/nist1a.cfm.
Pruitt, K.D., Tatusova, T., Brown, G.R. & Maglott, D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2012).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Bairoch, A. et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33, D154–D159 (2005).
Kersten, R.D. et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc. Natl. Acad. Sci. USA 110, E4407–E4416 (2013).
Guthals, A., Watrous, J.D., Dorrestein, P.C. & Bandeira, N. The spectral networks paradigm in high throughput mass spectrometry. Mol. Biosyst. 8, 2535–2544 (2012).
Mascuch, S.J. et al. Direct detection of fungal siderophores on bats with white-nose syndrome via fluorescence microscopy-guided ambient ionization mass spectrometry. PLoS One 10, e0119668 (2015).
Bandeira, N., Tsur, D., Frank, A. & Pevzner, P. Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. USA 104, 6140–6145 (2007).
Winnikoff, J.R., Glukhov, E., Watrous, J., Dorrestein, P.C. & Gerwick, W.H. Quantitative molecular networking to profile marine cyanobacterial metabolomes. J. Antibiot. (Tokyo) 67, 105–112 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Kildgaard, S. et al. Accurate dereplication of bioactive secondary metabolites from marine-derived fungi by UHPLC-DAD-QTOFMS and a MS/HRMS library. Mar. Drugs 12, 3681–3705 (2014).
Matsuda, F. et al. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 152, 566–578 (2010).
Haug, K. et al. MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 41, D781–D786 (2013).
Martens, L. et al. PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 (2005).
Uchida, K. & Zähner, H. Metabolic products of microorganisms 142. A new antibiotic derinamycin, inhibitor of DNA and RNA synthesis. J. Antibiot. (Tokyo) 28, 266–273 (1975).
Liu, W.-T. et al. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J. Antibiot. (Tokyo) 67, 99–104 (2014).
Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 49, 591–596 (1984).
Nonejuie, P., Burkart, M., Pogliano, K. & Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. USA 110, 16169–16174 (2013).
Lamsa, A., Liu, W.T., Dorrestein, P.C. & Pogliano, K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 84, 486–500 (2012).
Purves, K. et al. Using molecular networking for microbial secondary metabolite bioprospecting. Metabolites 6, 2 (2016).
Bertin, M.J. et al. Spongosine production by a Vibrio harveyi strain associated with the sponge Tectitethya crypta. J. Nat. Prod. 78, 493–499 (2015).
Boudreau, P.D. et al. Expanding the described metabolome of the marine cyanobacterium Moorea producens JHB through orthogonal natural products workflows. PLoS One 10, e0133297 (2015).
Kleigrewe, K. et al. Combining mass spectrometric metabolic profiling with genomic analysis: a powerful approach for discovering natural products from cyanobacteria. J. Nat. Prod. 78, 1671–1682 (2015).
Duncan, K.R. et al. Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem. Biol. 22, 460–471 (2015).
Vizcaino, M.I. & Crawford, J.M. The colibactin warhead crosslinks DNA. Nat. Chem. 7, 411–417 (2015).
Klitgaard, A., Nielsen, J.B., Frandsen, R.J.N., Andersen, M.R. & Nielsen, K.F. Combining stable isotope labeling and molecular networking for biosynthetic pathway characterization. Anal. Chem. 87, 6520–6526 (2015).
Anderton, C.R., Chu, R.K., Tolilc´, N., Creissen, A. & Paša-Tolic´, L. Utilizing a robotic sprayer for high lateral and mass resolution MALDI FT-ICR MSI of microbial cultures. J. Am. Soc. Mass Spectrom. 27, 556–559 (2016).
Liaimer, A. et al. Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc. Natl. Acad. Sci. USA 112, 1862–1867 (2015).
Liu, Y. et al. Diversity of aquatic pseudomonas species and their activity against the fish pathogenic oomycete saprolegnia. PLoS One 10, e0136241 (2015).
He, X. et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA 112, 244–249 (2015).
Cha, J.-Y. et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129 (2016).
Dührkop, K., Shen, H., Meusel, M., Rousu, J. & Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 112, 12580–12585 (2015).
Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Wishart, D.S. et al. HMDB: The human metabolome database. Nucleic Acids Res. 35, D521–D526 (2007).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2016).
Acknowledgements
This work was partially supported by US National Institutes of Health (NIH) grants 5P41GM103484-07, GM094802, AI095125, GM097509, S10RR029121, UL1RR031980, GM085770, U01TW0007401, and U01AI12316-01; N.B. was also partially supported as an Alfred P. Sloan Fellow. In addition, this work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), NIH, and the Department of Health and Human Services, under Contract Number HHSN272200800060C. V.V.P. is supported by the NIH grant K01 GM103809. L.M.S. is supported by NIH IRACDA K12 GM068524 award. T.L.-K. is supported by the United States–Israel Binational Agricultural Research and Development Fund Vaadia-BARD No. FI-494-13. C.P. is supported by Science without Borders Program from CNPq. A.M.C.R. is supported by São Paulo Research Foundation (FAPESP) grant#2014/01651-8, 2012/18031-7. K.K. was supported by a fellowship within the Postdoc-Programme of the German Academic Exchange Service (DAAD). M.C. was supported by a Deutsche Forschungsgemeinschaft (D.F.G.) postdoctoral fellowship. E.B. is supported by a Marie Curie IOF Fellowship within the 7th European Community Framework Program (FP7-PEOPLE-2011-IOF, grant number 301244-CYANOMIC). C.-C.L. was supported by a grant from the Ministry of Science and Technology of Taiwan (MOST103-2628-B-110-001-MY3). P.C. and B.Ø.P. were supported by the Novo Nordisk Foundation. Lixin Zhang and Xueting Liu are supported by the National Program on Key Basic Research Project (2013BC734000) and the National Natural Science Foundation of China (81102369 and 31125002). D.P. is supported by an INSA grant, Rennes. R.R.S. is supported by FAPESP grant#2014/01884-2. D.P.D. is supported by FAPESP grant#2014/18052-0. L.M.M. is supported by FAPESP grant#2013/16496-5. D.B.S. is supported by FAPESP grant#2012/18031-7. N.P.L. is supported by FAPESP (2014/50265-3), CAPES/PNPD, CNPq-PQ 480 306385/2011-2, and CNPq-INCT_if. E.A.G. is supported by the Notre Dame Chemistry-Biochemistry-Biology Interface (CBBI) program and NIH T32 GM075762. W.S. and J.S.M. are supported by grants from the National Institutes of Health 1R01DE023810-01 and 1R01GM095373. A.E. is supported by a grant from the NIH K99DE024543. C.F.M. and L.J. are supported by the Villum Foundation VKR023113, the Augustinus Foundation 13-4656, and the Aase & Ejnar Danielsens Foundation 10-001120. M.S.-C. was supported by UC MEXUS-CONACYT Collaborative Grant CN-12-552. M.F.T. was supported by NIH grant 1F32GM089044. Contributions by B.E.S. were supported by NSF grant DEB 1010816 and a Smithsonian Institution Grand Challenges Award. E.J.N.H. and J.P. are supported by the DFG (Forschergruppe 854) and by SNF grant IZLSZ3_149025. K.F.N. and A.K. are supported by the Danish Council for Independent Research, Technology, and Production Sciences (09-064967) and the Agilent Thought Leader Program. A.C.S. and R.S.B. were supported by NIH/NIAID U19-AI106772. B.T.M. and M.E. were supported under Department of Defense grant #W81XWH-13-1-0171. Contributions by O.B.V. and K.L.M. were supported by Oregon Sea Grant NA10OAR4170059/R/BT-48, NIH 5R21AI085540, and U01TW006634-06. E.E.C., A.M.S., and A.R.J. were supported by an NSF CAREER Award, a Pew Biomedical Scholar Award (E.E.C.), a Sloan Research Fellow Award (E.E.C.), the Research Corporation for Science Advancement (Cottrell Scholar Award; E.E.C.) and an Indiana University Quantitative Chemical Biology trainee fellowship (A.R.J.). M.M. was supported by the Danish Research Council for Technology and Production Science with Sapere Aude (116262). P.-M.A. was supported by FNS for fellowship on Subside (200020_146200). We thank V. Paul, R. Taylor, L. Aluwihare, F. Rohwer, B. Pullman, J. Fang, M. Overgaard, M. Katze, R.D. Smith, S.K. Mazmanian, W. Fenical, E. Macagno, X. He, and C. Neubauer for feedback and support for their laboratory personnel to contribute to the work. We thank B. Gust and co-workers at the University of Tuebingen for assisting us to obtain Streptomyces sp. DSM5940.
Author information
Authors and Affiliations
Contributions
Design and oversight of the project: P.C.D. and N.B. Algorithms: M.W. and N.B. Website: M.W., J.J.C. In-house library acquisition and analysis: V.V.P., L.M.S., N.G., A.J., D.-T.N., D.V., E.E., E.P., H.H., P.S., T.P., V.M. User-curated library acquisition and analysis: A.C.S., A.E., J.M., W.S., W.-T.L., M.J.M., V.V.P., L.M.S., N.G., R.A.Q., A.B., C.P., T.L.-K., A.M.C.R., A.M., M.C., K.R.D., K.K., E.C.O'N., B.S.M., E.B., E.G., D.D.N., S.J.M., P.D.B., X.L., L.Z., H.-U.H., C.F.M., L.J., D.P., S.T., E.A.G., M.S.-C., C.S., K.L.K., P.-M.A., R.G.L., R.S.B., P.R.J., M.F.T., S.J., B.E.S., L.M.M., DP.D., D.B.S., N.P.L., J.P., E.J.N.H., A.K., R.A.K., J.E.K., T.O.M., P.G.W., J.D., R.N., J.G., B.A., O.B.V., K.L.M., E.E.C., A.M.S., A.R.J., R.D.K., J.J.K., K.M.W., C.-C.H., M.M., C.-C.L., Y.-L.Y., A.V.M., C.B.L., D.J.G., F.R., H.M., J.-L.W., J.M., J.A., J.W., J.A.V., K.D., K.F.N., M.L., N.E., N.K., P. Pevzner, P. Phapale, R.J.D., R.B., R.M., R.G.G., T.A., T.H., T.N., V.A., W.H.G., Y.Z. Sample preparation, data generation, and website beta testing: A.E., W.T.L., M.J.M., V.V.P., L.M.S., N.G., R.A.Q., A.B., C.P., T.L.-K., A.M.C.R., A.M., D.J.F., M.C., J.J.C., N.B., P.C.D., E.C.O., E.B., E.G., D.D.N., S.J.M., P.D.B., X.L., L.Z., C.Z., C.F.M., R.R.S., E.A.G., M.S.-C., C.S., D.P., S.T., P.-M.A., R.G.L., B.E.S., L.M.M., J.P., E.J.N.H., D.T.-M., C.A.B.P., M.E., B.T.M., O.B.V., K.L.M., E.E.C., A.M.S., A.R.J., K.R.D. GNPS documentation: M.W., V.V.P., L.M.S., C.A.K., D.D.N., R.R.S., L.A.P. Genome sequencing, assembly and targeted amplification: Y.P., P.C., R.G.G., M.G., B.Ø.P., L.G. Stenothricin GNPS data analysis: W.-T.L., V.V.P., L.M.S., Y.P., P.C.D. NMR acquisition and analysis: B.M.D., P.D.B., L.M.S. Marfey's analysis: Y.P., P.D.B. Microbiology: Y.P., A.C.S., R.S.B. Peptidogenomics analysis: Y.P., R.D.K., P.C.D. Fluorescence Microscopy: Y.P., A.L., K.P. Writing of the paper: M.W., V.V.P., L.M.S., N.G., R.K., P.C.D., and N.B.
Ethics declarations
Competing interests
N.B. has an equity interest in Digital Proteomics, LLC, a company that may potentially benefit from the research results; Digital Proteomics, LLC, was not involved in any aspects of this research. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. E.E., E.P., H.H., L.V., and V.M. are employees of Sirenas MD. P.C.D. is on the advisory board for Sirenas MD. T.A. is the Scientific Director of SCiLS GmbH.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and 6–14, Supplementary Figures 1–20, Supplementary Notes 1–12 and Supplementary Methods (PDF 6806 kb)
Rights and permissions
About this article
Cite this article
Wang, M., Carver, J., Phelan, V. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34, 828–837 (2016). https://doi.org/10.1038/nbt.3597
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3597
This article is cited by
-
Apiospora arundinis, a panoply of carbohydrate-active enzymes and secondary metabolites
IMA Fungus (2024)
-
Genome sequencing and molecular networking analysis of the wild fungus Anthostomella pinea reveal its ability to produce a diverse range of secondary metabolites
Fungal Biology and Biotechnology (2024)
-
Mass spectrometry-guided isolation of thiodiketopiperazines from an EtOAc-extract of Setosphaeria rostrata culture medium and their anti-skin aging effects on TNF-α-induced human dermal fibroblasts
The Journal of Antibiotics (2024)
-
Fast mass spectrometry search and clustering of untargeted metabolomics data
Nature Biotechnology (2024)
-
nanoRAPIDS as an analytical pipeline for the discovery of novel bioactive metabolites in complex culture extracts at the nanoscale
Communications Chemistry (2024)