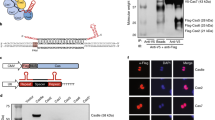
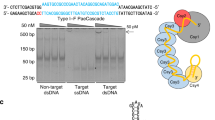
Abstract
We have developed a CRISPR-based method that uses catalytically active Cas9 and distinct single guide (sgRNA) constructs to knock out and activate different genes in the same cell. These sgRNAs, with 14- to 15-bp target sequences and MS2 binding loops, can activate gene expression using an active Streptococcus pyogenes Cas9 nuclease, without inducing double-stranded breaks. We use these 'dead RNAs' to perform orthogonal gene knockout and transcriptional activation in human cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
Change history
04 February 2016
In the version of this article initially published, when discussing the data in Figure 2b, on p. 1160, we wrote, “...targeting the same HBG1/2 promoter and found they had 32 and 55 perturbed transcripts....” This should have been “31 and 55 perturbed transcripts” as in the sentence in the figure legend discussing the same data. The error has been corrected in the HTML and PDF versions of the article.
References
Zalatan, J.G. et al. Cell 160, 339–350 (2015).
Esvelt, K.M. et al. Nat. Methods 10, 1116–1121 (2013).
Jinek, M. et al. Science 337, 816–821 (2012).
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Proc. Natl. Acad. Sci. USA 109, E2579–E2586 (2012).
Perez-Pinera, P. et al. Nat. Methods 10, 973–976 (2013).
Mali, P. et al. Nat. Biotechnol. 31, 833–838 (2013).
Maeder, M.L. et al. Nat. Methods 10, 977–979 (2013).
Konermann, S. et al. Nature 500, 472–476 (2013).
Gilbert, L.A. et al. Cell 154, 442–451 (2013).
Hilton, I.B. et al. Nat. Biotechnol. 33, 510–517 (2015).
Konermann, S. et al. Nature 517, 583–588 (2015).
Nishimasu, H. et al. Cell 156, 935–949 (2014).
Hsu, P.D. et al. Nat. Biotechnol. 31, 827–832 (2013).
Briner, A.E. et al. Mol. Cell 56, 333–339 (2014).
Wu, X. et al. Nat. Biotechnol. 32, 670–676 (2014).
Shalem, O. et al. Science 343, 84–87 (2014).
Platt, R.J. et al. Cell 159, 440–455 (2014).
Dow, L.E. et al. Nat. Biotechnol. 33, 390–394 (2015).
Fu, Y., Sander, J.D., Reyon, D., Cascio, V.M. & Joung, J.K. Nat. Biotechnol. 32, 279–284 (2014).
Kiani, S. et al. Nat. Methods. doi:10.1038/nmeth.3580 (7 September 2015).
Ran, F.A. et al. Cell 154, 1380–1389 (2013).
Li, B. & Dewey, C.N. BMC Bioinformatics 12, 323 (2011).
Acknowledgements
The authors thank K. Zheng and I. Slaymaker for their support and input. J.E.D. is supported by a Life Science Research Foundation post-doctoral fellowship of the Cystic Fibrosis Foundation. O.O.A. is supported by a Friends of the McGovern Institute Fellowship. J.S.G. is supported by a Department of Energy (DOE) Computational Science Graduate Fellowship. F.Z. is supported by the National Institute of Mental Health (NIMH) (1DP1-MH100706), the Poitras, Vallee, Simons, Paul G. Allen and New York Stem Cell Foundations, and David R. Cheng and Bob Metcalfe. The authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang laboratory website (http://www.genome-engineering.org/).
Author information
Authors and Affiliations
Contributions
J.E.D., O.O.A., F.Z. and S.K. conceived this study and designed the experiments. J.E.D., O.O.A., S.K., J.J. and J.S.G. performed experiments. J.E.D., O.O.A., F.Z. and S.K. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
J.E.D., O.O.A., F.Z. and S.K. have filed for intellectual property rights related to material described in this publication.
Integrated supplementary information
Supplementary Figure 1 dRNAs can mediate robust gene activation using an active Cas9.
Three different dRNAs targeting the HBG1 promoter region were designed. The length of the RNA targeting sequence was varied from 11nt to 20nt. HBG1 mRNA (normalized to GAPDH, and compared to cells transfected with GFP plasmid) was quantified, as well HBG1 indel frequency. In all cases, guides were designed with MS2 binding loops in the tetraloops and stem loop two11, and were co-transfected with active Cas9 and the MPH transcriptional activation complex. Average +/− SEM is plotted, N=2-3 replicates group.
Supplementary Figure 2 20bp dRNAs with mismatches at the 5′ end can activate transcription.
Four dRNAs were designed to target the HBG1 promoter region with a series of 5′ end mismatches (red). Target indel formation occurred consistently when sixteen or more nucleotides were matched to target DNA. Gene activation was observed with as few as 11 bp of homology to the target DNA. Average +/− SEM is plotted, N=2-3 replicates/ group.
Supplementary Figure 3 Transcriptome-wide mRNA profiles for ten different sgRNAs targeting HBG1/2
20nt sgRNAs with MS2 binding loops were co-transfected with dCas9 and the MPH complex. A previously published nuclease OT score13 did not significantly correlate with guide specificity. (In all cases, N=3 replicates / group).
Supplementary Figure 4 dRNAs activate target gene expression with active Cas9
A375 cells were transduced with lentivirus containing a dRNA. (a) Indel formation was measured at 0.6% and 0.05% for DNA sites targeted by ITGA9 and EGFR dRNAs, respectively. (b) ITGA9 and EGFR mRNA levels (normalized to GAPDH) were quantified. In all cases, average + / − SEM is plotted, and N=3 replicates / group.
Supplementary information
Supplementary Text
Supplementary Figures 1-4 and Supplementary Notes (PDF 2748 kb)
Rights and permissions
About this article
Cite this article
Dahlman, J., Abudayyeh, O., Joung, J. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol 33, 1159–1161 (2015). https://doi.org/10.1038/nbt.3390
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3390
This article is cited by
-
Doxycycline-dependent Cas9-expressing pig resources for conditional in vivo gene nullification and activation
Genome Biology (2023)
-
CasKAS: direct profiling of genome-wide dCas9 and Cas9 specificity using ssDNA mapping
Genome Biology (2023)
-
HideRNAs protect against CRISPR-Cas9 re-cutting after successful single base-pair gene editing
Scientific Reports (2022)
-
Make it a Combo
Nature Plants (2022)
-
Dead Cas(t) light on new life: CRISPRa-mediated reprogramming of somatic cells into neurons
Cellular and Molecular Life Sciences (2022)