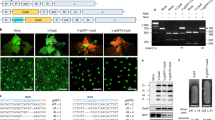
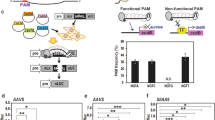
Abstract
Editing plant genomes without introducing foreign DNA into cells may alleviate regulatory concerns related to genetically modified plants. We transfected preassembled complexes of purified Cas9 protein and guide RNA into plant protoplasts of Arabidopsis thaliana, tobacco, lettuce and rice and achieved targeted mutagenesis in regenerated plants at frequencies of up to 46%. The targeted sites contained germline-transmissible small insertions or deletions that are indistinguishable from naturally occurring genetic variation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Li, J.F. et al. Nat. Biotechnol. 31, 688–691 (2013).
Shan, Q. et al. Nat. Biotechnol. 31, 686–688 (2013).
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J.D. & Kamoun, S. Nat. Biotechnol. 31, 691–693 (2013).
Kim, H. & Kim, J.S. Nat. Rev. Genet. 15, 321–334 (2014).
Jones, H.D. Nat. Plants 1, 14011 (2015).
Kim, S., Kim, D., Cho, S.W., Kim, J. & Kim, J.S. Genome Res. 24, 1012–1019 (2014).
Cho, S.W., Lee, J., Carroll, D., Kim, J.S. & Lee, J. Genetics 195, 1177–1180 (2013).
Sung, Y.H. et al. Genome Res. 24, 125–131 (2014).
Kim, J.M., Kim, D., Kim, S. & Kim, J.S. Nat. Commun. 5, 3157 (2014).
Kim, H.J., Lee, H.J., Kim, H., Cho, S.W. & Kim, J.S. Genome Res. 19, 1279–1288 (2009).
Lee, H.J., Kim, E. & Kim, J.S. Genome Res. 20, 81–89 (2010).
Bae, S., Park, J. & Kim, J.S. Bioinformatics 30, 1473–1475 (2014).
Cho, S.W. et al. Genome Res. 24, 132–141 (2014).
Cho, S.W., Kim, S., Kim, J.M. & Kim, J.S. Nat. Biotechnol. 31, 230–232 (2013).
Choe, S. et al. Plant Physiol. 130, 1506–1515 (2002).
Kim, D. et al. Nat. Methods 12, 237–243 (2015).
Kanchiswamy, C.N., Malnoy, M., Velasco, R., Kim, J.S. & Viola, R. Trends Biotechnol. 33, 489–491 (2015).
Yoo, S.D., Cho, Y.H. & Sheen, J. Nat. Protoc. 2, 1565–1572 (2007).
Zhang, Y. et al. Plant Methods 7, 30 (2011).
Lelivelt, C.L. et al. Plant Mol. Biol. 58, 763–774 (2005).
Frearson, E.M., Power, J.B. & Cocking, E.C. Dev. Biol. 33, 130–137 (1973).
Menczel, L., Nagy, F., Kiss, Z.R. & Maliga, P. Theor. Appl. Genet. 59, 191–195 (1981).
Gamborg, O.L., Miller, R.A. & Ojima, K. Exp. Cell Res. 50, 151–158 (1968).
Acknowledgements
This work was supported in part by grants from the Institute for Basic Science (IBS-R021-D1) and the Next-Generation BioGreen21 Program (PJ01104501 to S.C. and PJ01104502 to S.I.K.).
Author information
Authors and Affiliations
Contributions
J.-S.K. and S.C. supervised the research. J.W.W., S.I.K. and C.C. carried out plant regeneration. J.K., S.W.C. H.K., S.-G.K. and S.-T.K. performed mutation analysis.
Corresponding authors
Ethics declarations
Competing interests
J.-S.K. and S.C. are co-inventors on a patent application covering the genome editing method described in this manuscript.
Integrated supplementary information
Supplementary Figure 1 Analysis of off-target effects.
Mutation frequencies at on-target and potential off-target sites of the PHYB and BRI1 gene-specific sgRNAs were measured by targeted deep sequencing. About ~80,000 paired-end reads per site were obtained to calculate the indel rate.
Supplementary Figure 2 Partial nucleotide and amino acid sequences of LsBIN2.
Underscored and boxed letters represent the sequences corresponding to degenerate primers and sgRNA, respectively.
Supplementary Figure 3 Regeneration of plantlets from RGEN RNP-transfected protoplast in L. sativa.
Protoplast division, callus formation and shoot regeneration from RGEN RNP-transfected protoplasts in the lettuce. (a) Cell division after 5 days of protoplast culture (Bar = 100 μm). (b) A multicellular colony of protoplast (Bar = 100 μm). (c) Agarose-embedded colonies after 4 weeks of protoplast culture. (d) Callus formation from protoplast-derived colonies (e,f) Organogenesis and regenerated shoots from protoplast-derived calli (bar = 5 mm).
Supplementary Figure 4 Targeted deep sequencing of mutant calli.
Genotypes of the mutant calli were confirmed by Illumina Miseq. Sequence of each allele and the number of sequencing reads were analyzed. (A1), allele1. (A2), allele2.
Supplementary Figure 5 Plant regeneration from RGEN RNP-transfected protoplasts in L. sativa.
(a-c) Organogenesis and shoot formation from protoplast-derived calli; wild type (#28), bi-allelic/heterozygote (#24), bi-allelic/homozygote (#30). (d) In vitro shoot proliferation and development. (e) Elongation and growth of shoots in MS culture medium free of PGR. (f) Root induction onto elongated shoots. (g) Acclimatization of plantlets. (h,i) Regenerated whole plants.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 1173 kb)
Rights and permissions
About this article
Cite this article
Woo, J., Kim, J., Kwon, S. et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33, 1162–1164 (2015). https://doi.org/10.1038/nbt.3389
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3389
This article is cited by
-
Targeted gene knockout via CRISPR/Cas9: precise genome editing in eggplant (Solanum melongena) through phytoene desaturase gene disruption
Journal of Crop Science and Biotechnology (2024)
-
The applications of CRISPR/Cas-mediated microRNA and lncRNA editing in plant biology: shaping the future of plant non-coding RNA research
Planta (2024)
-
CRISPR-edited plants by grafting
Nature Biotechnology (2023)
-
Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks
Nature Biotechnology (2023)
-
Transgene-free genome editing of vegetatively propagated and perennial plant species in the T0 generation via a co-editing strategy
Nature Plants (2023)