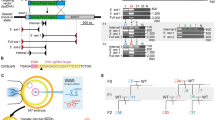
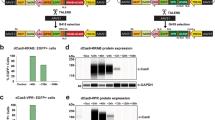
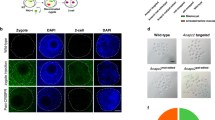
Abstract
Identification of genes influencing a phenotype of interest is frequently achieved through genetic screening by RNA interference (RNAi) or knockouts. However, RNAi may only achieve partial depletion of gene activity, and knockout-based screens are difficult in diploid mammalian cells. Here we took advantage of the efficiency and high throughput of genome editing based on type II, clustered, regularly interspaced, short palindromic repeats (CRISPR)–CRISPR-associated (Cas) systems to introduce genome-wide targeted mutations in mouse embryonic stem cells (ESCs). We designed 87,897 guide RNAs (gRNAs) targeting 19,150 mouse protein-coding genes and used a lentiviral vector to express these gRNAs in ESCs that constitutively express Cas9. Screening the resulting ESC mutant libraries for resistance to either Clostridium septicum alpha-toxin or 6-thioguanine identified 27 known and 4 previously unknown genes implicated in these phenotypes. Our results demonstrate the potential for efficient loss-of-function screening using the CRISPR-Cas9 system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Forsburg, S.L. The art and design of genetic screens: yeast. Nat. Rev. Genet. 2, 659–668 (2001).
Jorgensen, E.M. & Mango, S.E. The art and design of genetic screens: Caenohabditis elegans. Nat. Rev. Genet. 3, 356–369 (2002).
Boutros, M. & Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9, 554–566 (2008).
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Iorns, E., Lord, C.J., Turner, N. & Ashworth, A. Utilizing RNA interference to enhance cancer drug discovery. Nat. Rev. Drug Discov. 6, 556–568 (2007).
Carette, J.E. et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 (2009).
Carette, J.E. et al. Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29, 542–546 (2011).
Carette, J.E. et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 (2011).
Leeb, M. & Wutz, A. Derivation of haploid embryonic stem cells from mouse embryos. Nature 479, 131–134 (2011).
Yang, H. et al. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell 149, 605–617 (2012).
Elling, U. et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell 9, 563–574 (2011).
Yang, H. et al. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 23, 1187–1200 (2013).
Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang, H.S. & Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Joung, J.K. & Sander, J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Cho, S.W., Kim, S., Kim, J.M. & Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 230–232 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Takeda, J. et al. Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell 73, 703–711 (1993).
Kinoshita, T., Fujita, M. & Maeda, Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J. Biochem. 144, 287–294 (2008).
Gordon, V.M. et al. Clostridium septicum alpha toxin uses glycosylphosphatidylinositol-anchored protein receptors. J. Biol. Chem. 274, 27274–27280 (1999).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Ellis, J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 16, 1241–1246 (2005).
Bennardo, N., Cheng, A., Huang, N. & Stark, J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 (2008).
Nichols, J. et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 (1998).
Mitsui, K. et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 (2003).
Hakem, R. et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85, 1009–1023 (1996).
Tsuzuki, T. et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 93, 6236–6240 (1996).
Luo, J. et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 (2009).
Agaisse, H. et al. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309, 1248–1251 (2005).
Karlas, A. et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 (2010).
Ran, F.A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Pattanayak, V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843 (2013).
Cadinanos, J. & Bradley, A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 35, e87 (2007).
Yusa, K., Rad, R., Takeda, J. & Bradley, A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods 6, 363–369 (2009).
Carey, B.W. et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA 106, 157–162 (2009).
Subach, O.M. et al. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15, 1116–1124 (2008).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Pettitt, S.J. et al. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods 6, 493–495 (2009).
Wang, W., Bradley, A. & Huang, Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 19, 667–673 (2009).
Abuin, A., Zhang, H. & Bradley, A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol. Cell. Biol. 20, 149–157 (2000).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Quail, M.A. et al. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 9, 10–11 (2012).
Huang, da W. et al. DAVID gene ID conversion tool. Bioinformation 2, 428–430 (2008).
Acknowledgements
We thank A. Bradley for comments on the manuscript, B. Ng and W. Cheng for the flow cytometry analyses, and the Sanger Institute DNA pipeline for the sequence analyses. We also thank J. Takeda and T. Kinoshita for providing us alpha-toxin and the cDNA expression vectors, respectively. Y.L. is supported by the Wellcome Trust PhD program. M.D.C.V.-H. is supported by the Cancer Research UK and Wellcome Trust PhD program. This work was supported by Wellcome Trust (WT077187). The mouse CRISPR library is available through Addgene. The plasmid DNAs are available at the Wellcome Trust Sanger Institute Archives (http://www.sanger.ac.uk/technology/clonerequests/).
Author information
Authors and Affiliations
Contributions
K.Y. conceived the research and wrote the manuscript with comments from all authors. H.K.-Y., E.-P.T. and K.Y. performed the experiments. Y.L. and M.D.C.V.-H. performed the bioinformatics analyses.
Corresponding author
Ethics declarations
Competing interests
K.Y. and Y.L. filed a patent application based on the results reported in this paper.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–19 and Supplementary Tables 3–7 (PDF 2520 kb)
Supplementary Table 1
Off-target cleavage analyses of the gRNA targeting Site 2 of the Piga gene (with no bulge) (XLSX 101 kb)
Supplementary Table 2
Off-target cleavage analyses of the gRNA targeting Site 2 of the Piga gene (with bulges) (XLSX 157 kb)
Supplementary Table 8
List of shRNAs and oligo seqeunces (XLSX 10 kb)
Supplementary Table 9
List of genes, gRNA target sequences and oligonucleotide sequences used in this study. (XLSX 18 kb)
Supplementary Data 1
(XLSX 35502 kb)
Supplementary Data 2
(XLSX 6296 kb)
Supplementary Data 3
A full list of potential off-target sites (with NGG PAM) of the gRNA targeting site 2 of the Piga gene. (XLSX 338 kb)
Supplementary Data 4
A full list of potential off-target sites (with NAG PAM) of the gRNA targeting Site 2 of the Piga gene. (XLSX 455 kb)
Supplementary Data 5
A full list of potential off-target sites (with bulges) of the gRNA targeting site 2 of the Piga gene. (XLSX 76 kb)
Source data
Rights and permissions
About this article
Cite this article
Koike-Yusa, H., Li, Y., Tan, EP. et al. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32, 267–273 (2014). https://doi.org/10.1038/nbt.2800
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2800
This article is cited by
-
Genome-wide CRISPR screening identifies a role for ARRDC3 in TRP53-mediated responses
Cell Death & Differentiation (2024)
-
Genetic determinants of micronucleus formation in vivo
Nature (2024)
-
A multi-omics integrative analysis based on CRISPR screens re-defines the pluripotency regulatory network in ESCs
Communications Biology (2023)
-
Deep sampling of gRNA in the human genome and deep-learning-informed prediction of gRNA activities
Cell Discovery (2023)
-
Mitochondrial E3 ubiquitin ligase MARCHF5 controls BAK apoptotic activity independently of BH3-only proteins
Cell Death & Differentiation (2023)