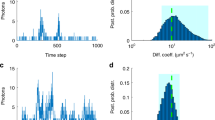
Abstract
Diffusion processes and local dynamic equilibria inside cells lead to nonuniform spatial distributions of molecules, which are essential for processes such as nuclear organization and signaling in cell division, differentiation and migration1. To understand these mechanisms, spatially resolved quantitative measurements of protein abundance, mobilities and interactions are needed, but current methods have limited capabilities to study dynamic parameters. Here we describe a microscope based on light-sheet illumination2 that allows massively parallel fluorescence correlation spectroscopy (FCS)3 measurements and use it to visualize the diffusion and interactions of proteins in mammalian cells and in isolated fly tissue. Imaging the mobility of heterochromatin protein HP1α (ref. 4) in cell nuclei we could provide high-resolution diffusion maps that reveal euchromatin areas with heterochromatin-like HP1α-chromatin interactions. We expect that FCS imaging will become a useful method for the precise characterization of cellular reaction-diffusion processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kinkhabwala, A. & Bastiaens, P.L.H. Spatial aspects of intracellular information processing. Curr. Opin. Genet. Dev. 20, 31–40 (2010).
Huisken, J. & Stainier, D.Y.R. Selective plane illumination microscopy techniques in developmental biology. Development 136, 1963–1975 (2009).
Elson, E.L. & Magde, D. Fluorescence correlation spectroscopy. I Conceptual basis and theory. Biopolymers 13, 1–27 (1974).
Cheutin, T. et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 (2003).
Bacia, K., Kim, S.A. & Schwille, P. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods 3, 83–89 (2006).
Kim, S.A., Heinze, K.G. & Schwille, P. Fluorescence correlation spectroscopy in living cells. Nat. Methods 4, 963–973 (2007).
Maeder, C.I. et al. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nat. Cell Biol. 9, 1319–1326 (2007).
Slaughter, B.D., Schwartz, J.W. & Li, R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc. Natl. Acad. Sci. USA 104, 20320–20325 (2007).
Schmidt, U. et al. Assembly and mobility of exon-exon junction complexes in living cells. RNA 15, 862–876 (2009).
Kawai-Noma, S. et al. Dynamics of yeast prion aggregates in single living cells. Genes Cells 11, 1085–1096 (2006).
Yu, S.R. et al. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature 461, 533–536 (2009).
Wachsmuth, M., Waldeck, W. & Langowski, J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298, 677–689 (2000).
Dertinger, T. et al. Two-focus fluorescence correlation spectroscopy: a new tool for accurate and absolute diffusion measurements. ChemPhysChem 8, 433–443 (2007).
Digman, M.A. et al. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 89, 1317–1327 (2005).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E.H.K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Wohland, T., Shi, X., Sankaran, J. & Stelzer, E.H. Single plane illumination fluorescence correlation spectroscopy (SPIM-FCS) probes inhomogeneous three-dimensional environments. Opt. Express 18, 10627–10641 (2010).
Kannan, B. et al. Spatially resolved total internal reflection fluorescence correlation microscopy using an electron multiplying charge-coupled device camera. Anal. Chem. 79, 4463–4470 (2007).
Heuvelman, G., Erdel, F., Wachsmuth, M. & Rippe, K. Analysis of protein mobilities and interactions in living cells by multifocal fluorescence fluctuation microscopy. Eur. Biophys. J. 38, 813–828 (2009).
Needleman, D.J., Xu, Y. & Mitchison, T.J. Pin-hole array correlation imaging: highly parallel fluorescence correlation spectroscopy. Biophys. J. 96, 5050–5059 (2009).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Gurdon, J.B. & Bourillot, P.Y. Morphogen gradient interpretation. Nature 413, 797–803 (2001).
Dinant, C. & Luijsterburg, M.S. The emerging role of HP1 in the DNA damage response. Mol. Cell Biol. 29, 6335–6340 (2009).
Kwon, S.H. & Workman, J.L. The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol. Cells 26, 217–227 (2008).
Grewal, S.I. & Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 301, 798–802 (2003).
Hediger, F. & Gasser, S.M. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 16, 143–150 (2006).
Müller, K.P. et al. Multiscale analysis of dynamics and interactions of heterochromatin protein 1 by fluorescence fluctuation microscopy. Biophys. J. 97, 2876–2885 (2009).
Schotta, G. et al. A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004).
Grewal, S.I. & Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007).
Holekamp, T.F., Turaga, D. & Holy, T.E. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron 57, 661–672 (2008).
Keller, P.J. et al. Fast, high-contrast imaging of animal development with scanned light-sheet-based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Planchon, T.A. et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 (2011).
Ritter, J.G., Veith, R., Veenendaal, A., Siebrasse, J.P. & Kubitscheck, U. Light-sheet microscopy for single molecule tracking in living tissue. PLoS ONE 5, e11639 (2010).
Bestvater, F. et al. EMCCD-based spectrally resolved fluorescence correlation spectroscopy. Opt. Express 18, 23818–23828 (2010).
Im, K.B. et al. Two-photon spectral imaging with high temporal and spectral resolution. Opt. Express 18, 26905–26914 (2010).
Gregor, I., Patra, D. & Enderlein, J. Optical saturation in fluorescence correlation spectroscopy under continuous-wave and pulsed excitation. ChemPhysChem 6, 164–170 (2005).
Gröner, N., Capoulade, J., Cremer, C. & Wachsmuth, M. Measuring and imaging diffusion with multiple scan speed image correlation spectroscopy. Opt. Express 18, 21225–21237 (2010).
Huh, W.K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Acknowledgements
We thank the mechanical and the electronics workshop of the European Molecular Biology Laboratory (EMBL) for custom hardware, M. Meurer for yeast cell culture, D. Holzer for mammalian cell culture, T. Weimbs for providing the MDCK II cells, A. Ephrussi for providing the flies expressing Ubi-GFP-NLS and K. Rippe for providing 3T3 cells expressing HP1α-EGFP. We would like to thank Leica Microsystems as well as R. Pepperkok, J. Ellenberg and the Advanced Light Microscopy Facility of EMBL for support. A. Aulehla, P. Keller and A. Khmelinskii are kindly acknowledged for helpful comments, as are many other colleagues for discussions. L.H. was supported by the center for modeling and simulation in the biosciences (BioMS). We are grateful for financial support from EMBL and from the EpiSys project within the BMBF SysTec program (grant no. 0315502C to M.W.).
Author information
Authors and Affiliations
Contributions
M.K. and M.W. conceived the research. J.C. implemented the light-pad microscope. J.C. and M.W. conducted the yeast and mammalian work. J.C., M.W. and L.H. conducted the Drosophila wing disc work. J.C., M.W. and M.K. analyzed the data and wrote the manuscript. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.C., M.K. and M.W. are named inventors on a patent application on technologies described in this manuscript.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1, Supplementary Results and Supplementary Figures 1–8 (PDF 2661 kb)
Supplementary Video S1
z-stack of images of yeast cells expressing Pma1-GFP acquired with the light-pad microscope (MOV 784 kb)
Supplementary Video S2
1D-FCS recording of 20 nm fluorescent beads diffusing in water (MOV 5291 kb)
Supplementary Video S3
2D-FCS recording of 20 nm fluorescent beads diffusing in water (MOV 1460 kb)
Supplementary Video S4
20 nm fluorescent beads diffusing in water recorded with the imaging camera showing Brownian motion and convective flow (MOV 1684 kb)
Supplementary Video S5
3D reconstruction of a Drosophila larva wing imaginal disc expressing GFP-NLS acquired with the light-pad microscope (MOV 1734 kb)
Rights and permissions
About this article
Cite this article
Capoulade, J., Wachsmuth, M., Hufnagel, L. et al. Quantitative fluorescence imaging of protein diffusion and interaction in living cells. Nat Biotechnol 29, 835–839 (2011). https://doi.org/10.1038/nbt.1928
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1928
This article is cited by
-
Light sheet fluorescence microscopy
Nature Reviews Methods Primers (2021)
-
Quantitative mapping of fluorescently tagged cellular proteins using FCS-calibrated four-dimensional imaging
Nature Protocols (2018)
-
Comprehensive correlation analysis for super-resolution dynamic fingerprinting of cellular compartments using the Zeiss Airyscan detector
Nature Communications (2018)
-
Local raster image correlation spectroscopy generates high-resolution intracellular diffusion maps
Communications Biology (2018)