Abstract
Oncogene-induced DNA replication stress contributes critically to the genomic instability that is present in cancer1,2,3,4. However, elucidating how oncogenes deregulate DNA replication has been impeded by difficulty in mapping replication initiation sites on the human genome. Here, using a sensitive assay to monitor nascent DNA synthesis in early S phase, we identified thousands of replication initiation sites in cells before and after induction of the oncogenes CCNE1 and MYC. Remarkably, both oncogenes induced firing of a novel set of DNA replication origins that mapped within highly transcribed genes. These ectopic origins were normally suppressed by transcription during G1, but precocious entry into S phase, before all genic regions had been transcribed, allowed firing of origins within genes in cells with activated oncogenes. Forks from oncogene-induced origins were prone to collapse, as a result of conflicts between replication and transcription, and were associated with DNA double-stranded break formation and chromosomal rearrangement breakpoints both in our experimental system and in a large cohort of human cancers. Thus, firing of intragenic origins caused by premature S phase entry represents a mechanism of oncogene-induced DNA replication stress that is relevant for genomic instability in human cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Halazonetis, T. D., Gorgoulis, V. G. & Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355 (2008)
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010)
Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010)
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013)
Ekholm-Reed, S. et al. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165, 789–800 (2004)
Jones, R. M. et al. Increased replication initiation and conflicts with transcription underlie cyclin E-induced replication stress. Oncogene 32, 3744–3753 (2013)
Beck, H. et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32, 4226–4236 (2012)
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006)
Bester, A. C. et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145, 435–446 (2011)
Aird, K. M. et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Reports 3, 1252–1265 (2013)
Toledo, L. I. et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013)
Kotsantis, P. et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat. Commun. 7, 13087 (2016)
Bartkova, J. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006)
Costantino, L. et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science 343, 88–91 (2014)
Maya-Mendoza, A. et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 9, 601–616 (2015)
Resnitzky, D., Gossen, M., Bujard, H. & Reed, S. I. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14, 1669–1679 (1994)
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003)
MacAlpine, D. M., Rodríguez, H. K. & Bell, S. P. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 18, 3094–3105 (2004)
Karnani, N. & Dutta, A. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 25, 621–633 (2011)
Hansen, R. S. et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–144 (2010)
Sasaki, T. et al. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol. Cell. Biol. 26, 1051–1062 (2006)
Powell, S. K. et al. Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 34, 531–543 (2015)
Hu, J. et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protocols 11, 853–871 (2016)
Wilson, T. E. et al. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 25, 189–200 (2015)
Helmrich, A., Ballarino, M. & Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 44, 966–977 (2011)
Letessier, A. et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 (2011)
Barlow, J. H. et al. Identification of early replicating fragile sites that contribute to genome instability. Cell 152, 620–632 (2013)
Prado, F. & Aguilera, A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 24, 1267–1276 (2005)
Petryk, N. et al. Replication landscape of the human genome. Nat. Commun. 7, 10208 (2016)
Martin, M. M. et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 21, 1822–1832 (2011)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Acknowledgements
We thank U. Schibler, R. Pillai, M. Docquier and present and past laboratory members for helpful discussions; J. Bartek for the U2OS cells inducibly overexpressing cyclin E; M. Eilers for the U2OS MycER cells; R. Beroukhim and S. Schumacher for access to and help with TCGA cancer data sets; and N. Roggli for help with the graphics scripts and the Flow Cytometry and Genomics platforms of the University of Geneva. This work was supported by grants from the European Commission (ONIDDAC) and the Swiss Science National Foundation.
Author information
Authors and Affiliations
Contributions
T.D.H. and M.M. designed the experiments and wrote the paper; M.M. performed the experiments; T.D.H. wrote the computer scripts and performed the bioinformatic analyses with contributions from M.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Experimental setup to study S phase entry and DNA replication initiation.
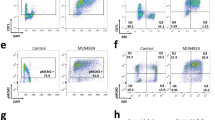
a, Cyclin E protein levels, as determined by immunoblotting, in NE and OE cells (2.5 days after tetracycline withdrawal). Actinin serves as a loading control. This is a representative example of more than ten independent replicates. b, Experimental outline of the protocol used to monitor S phase entry by flow cytometry and of the EdUseq protocols: EdUseq-HU, EdUseq-noHU and EdUseq-HU/release. *EdU was added 30 min before harvesting the cells. c, Flow cytometry profiles of NE and OE cells after mitotic shake-off (0 h) and 14 and 10 h later, respectively, after the cells had been released in medium containing hydroxyurea and EdU. 2C and 4C, DNA content of G1 and G2 cells, respectively. This is a representative example of more than ten independent replicates. d, DNA content versus EdU incorporation flow cytometry plots of NE and OE cells. EdU-positive NE and OE cells were gated blue and red, respectively. 2C and 4C, DNA content of G1 and G2 cells, respectively. The gating strategy for these data is shown in Supplementary Fig. 1.
Extended Data Figure 2 Identification of replication timing domains by REPLIseq.
a, Experimental outline of the REPLIseq protocol and FACS profiles of cells with normal levels of cyclin E (NE) and cells overexpressing cyclin E (OE) for 1, 2 or 7 days (1 d, 2 d or 7 d). Cells were sorted according to DNA content into early (blue), mid (green) and late (yellow) S phase fractions. 2C and 4C, DNA content of G1 and G2 cells, respectively. b, Assignment of RT domains. The fractions of the genome that were replicated in early, mid or late S phase were determined on the basis of the REPLIseq profiles of the NE and 2 d OE cells. c, Distribution of early, mid and late replication timing bins in 2 d OE cells according to their replication timing in NE cells. d, Comparison of the origin firing profile determined by EdUseq-HU (Fig. 1b) and the early S replication profile determined by REPLIseq in NE cells. RT domains and Ge/iG regions are as in Fig. 1b. Bin resolution, 10 kb; ruler scale, 100 kb. e, REPLIseq profiles of the first 10 Mb of chromosome 7 of NE or OE 1 d, 2 d or 7 d cells. Profiles are shown separately for the cells in early, mid and late S phase. RT domains and Ge/iG regions are as in Fig. 1b. Bin resolution, 10 kb; ruler scale, 100 kb.
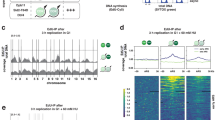
Extended Data Figure 3 Further characterization of replication origins.
a, Adjusted average sigma values at 1 kb resolution around 1,828 origins refined by performing the EdUseq-HU protocol in the additional presence of mimosine or aphidicolin (light and dark grey, respectively), compared to OE and NE cells treated only with hydroxyurea (pink and blue, respectively). b, Distribution of the subset of origins refined at 1 kb resolution (1,828 origins) relative to origin type, as determined by the original 10-kb resolution assignment (CN, Oi2 or Oi). c, Distribution of constitutive and oncogene-induced origins, refined at 1 kb resolution (1,828 origins), according to RT domains (E, early; M, mid; L, late S phase). d, Distribution of constitutive and oncogene-induced origins, refined at 1 kb resolution (1,828 origins), according to gene annotation in all RT domains (all-RT) or only in the early S phase RT domains (E-RT). e, Transcription (EUseq) levels (median) in NE (light grey) and OE (dark grey) cells at sites of constitutive and oncogene-induced origins, refined at 1 kb resolution, at various time points after mitotic shake-off. Origins mapping to genic or intergenic genomic bins were plotted separately. f, Scatter plots of EdUseq-noHU sigma values (log2) at all origins (CN, purple; Oi2, pink; Oi, red) at 10 kb resolution for NE versus OE cells not treated with hydroxyurea (Fig. 1h). EdU was present during the indicated time periods.
Extended Data Figure 4 Further characterization of transcription profiles and effects of DRB.
a, Genome-wide comparison (all genomic bins) of newly synthesized transcripts (EUseq) in NE versus OE cells at the indicated time points after mitotic shake-off. The two genomic bins mapping to CCNE1 are red. b, List of early and mid S large genes along which replication initiation and transcription profiles were plotted in Fig. 2d. For each gene, the association with a common fragile site (CFS) is indicated24. c, Inhibition of transcription by DRB. EU incorporation was monitored by fluorescence microscopy in control (no DRB), DRB-treated (9 h), and DRB-treated (9 h) and released (5 h) cells. The nuclei of the cells were counterstained with DAPI. d, DNA content versus EdU incorporation flow cytometry plots of NE cells 14 h after mitotic shake-off. The cells were treated with DRB for the indicated times. EdU-positive NE cells were gated in blue. 2C and 4C, DNA content of G1 and G2 cells, respectively. e, Newly synthesized transcript profiles (EUseq) at a representative genomic region in NE cells treated with DRB for 9 h after mitotic shake-off and then released for 30 or 120 min (release 30 min, dark grey; release 120 min, light grey; overlap: colour; direction of transcription: green, forward (forw); red, reverse (rev); yellow, bidirectional (bidir)). The red arrow indicates the transcription of the gene harbouring oncogene-induced origins at our example locus on chromosome 7 and the green arrow indicates another large gene in this locus. RT domain and Ge/iG annotations are as in Fig. 1b. f, Average transcription (EUseq, log2rσ) in NE cells treated with DRB for 9 h after mitotic-shake-off and then released for 30, 120 or 240 min, along the length of large genes (>0.35 Mb for early S and >0.65 Mb for mid S genes). The genes are grouped according to replication timing (RT; early, mid S) and level of transcription (high Tx, upper tercile; mid Tx, middle tercile). # Ge, number of genes averaged at each position.
Extended Data Figure 5 Accelerated entry into S phase and firing of novel intragenic origins upon Myc activation.
a, Myc activation (3 days after adding 4-OHT), as determined by immunfluorescence, in cells with non-induced (NM) and induced (OM) Myc activity. Nuclei counterstained with DAPI. Representative images from two independent experiments are shown. b, Quantification of EdU-positive cells at different time points after mitotic shake-off. Means and s.d.s were calculated from three independent experiments; grey symbols, individual data points. c, Replication initiation (EdUseq-HU) profiles at a representative genomic region in OM and NM cells, collected at the indicated times after mitotic shake-off. Peak heights are represented as sigma values (σ). RT domains and gene annotations are as in Fig. 1b. Bin resolution, 10 kb; ruler scale, 100 kb. d, Classification of constitutive and oncogene-induced origins based on relative height ratios in OM versus NM cells (OM:NM). e, Scatter plots of EdUseq-HU σ values at origins (CN, purple; Oi2, pink; Oi, red) in NM versus OM cells at the indicated time points after mitotic shake-off. f, Distribution of CN, Oi2 and Oi origins in OM and NM cells according to RT domains (E, early; M, mid; L, late S phase). g, Distribution of CN, Oi2 and Oi origins in OM and NM cells according to gene annotation in all replication timing domains (all-RT) or only in the early S phase replicating domains (E-RT). h, Relative adjusted sigma ratios of replication origins identified in NE, NM, OE or OM cells. Left, number of origins identified in NE or NM cells grouped according to their relative height ratios between these two cell lines. Right, number of Oi origins identified in OE or OM cells grouped according to their level of induction relative to the NE and NM cells, respectively. i, Newly synthesized transcript profiles (EUseq) at a representative genomic region in OM and NM cells 10 and 14 h after mitotic shake-off, respectively (NM: light grey; OM: dark grey; overlap: green, forward (forw); red, reverse (rev); yellow, bidirectional (bidir) direction of transcription). RT domains and gene annotations are as in c. j, Genome-wide comparison (all genomic bins) of transcription in OM versus NM cells 10 and 14 h after mitotic shake-off, respectively. k, Median transcription (EUseq) levels in NM (light grey) and OM (dark grey) cells at constitutive and oncogene-induced origins mapping to genic (Ge) or intergenic (iG) genomic bins at 14 and 10 h after mitotic shake-off, respectively.
Extended Data Figure 6 S phase entry and replication initiation profiles of HeLa and RPE1 cells.
a, Percentage of EdU-positive HeLa and RPE1 cells at different time points after mitotic shake-off (0 h). Means and individual data points are shown from two independent experiments. b, Replication initiation (EdUseq-HU) profiles at a representative genomic region in HeLa and RPE1 cells at the indicated time points after mitotic shake-off. The profile of NE U2OS cells (blue) serves as reference. c, Scatter plots of EdUseq-HU σ values (log2) at all origins (CN, purple; Oi2, pink; Oi, red) in HeLa and RPE1 cells versus NE U2OS cells at the indicated time points after mitotic shake-off.
Extended Data Figure 7 Replication initiation and transcription profiles at selected genomic loci.
a, Replication initiation (EdUseq-HU) profiles at three genomic loci in different cells lines, from top to bottom: OE versus NE cells, collected 6 and 14 h after mitotic shake-off, respectively; OM versus NM cells, collected 6 and 14 h after mitotic shake-off, respectively; HeLa cells collected at 6 versus 14 h after mitotic shake-off; and RPE1 cells collected 14 h after mitotic shake-off. Peak heights are represented as sigma values. RT domains and gene annotations are as in Fig. 1b. Bin resolution, 10 kb; ruler scale, 100 kb. The green arrows indicate the direction of transcription of the example gene of each locus harbouring oncogene-induced origins. b, Replication initiation (EdUseq-HU) profiles of control (noDRB) and DRB-treated (0–9 h) NE cells collected 14 h after mitotic shake-off. The same genomic loci as in a are shown, focusing on the genes harbouring the oncogene-induced origins. RT domains and gene annotations are as in a. c, Newly synthesized transcript profiles (EUseq) of NE cells, 2 and 14 h after mitotic shake-off (2 h, light green; 14 h, grey; overlap, dark green) shown only for the example genes harbouring the oncogene-induced origins shown in a (green arrows). RT domains and gene annotations are as in a.
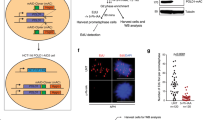
Extended Data Figure 8 Fork collapse at Oi origins induced in NE cells by inhibiting transcription in early G1.
a, Replication initiation (EdUseq-HU, 14 h hydroxyurea block) and fork progression (EdUseq-HU/release 60 min) profiles at a representative genomic region in NE U2OS cells treated or not with DRB during the first 7 h of G1. RT domains and gene annotations are as in Fig. 1b. Bin resolution, 10 kb; ruler scale, 100 kb. b, Average fork progression (no release and 60 min release) at constitutive and oncogene-induced origins located in highly transcribed regions in control and DRB treated (first 7 h of G1 phase) NE cells.
Extended Data Figure 9 Association of Oi origins with genomic rearrangements and replication timing profiles of cancer rearrangement breakpoints.
a, Mapping of translocations (Transloc; n = 27,364) identified by LAM-HTGTS to genomic regions replicated from Oi origins (OiRDs) with the analysis restricted to the early S replicating domains. The fraction of translocations mapping to OiRDs is shown for non-transcribed (0), low (Lo), medium (Me) and highly (Hi) transcribed genomic bins, as well as for all early S replicating bins. Statistical comparisons, using random permutation analysis, are between the NE (blue) and OE (pink) samples. The distribution of OiRDs in the genome (grey) is shown as a reference. b, Mapping of genomic rearrangement (Rearr; n = 136) breakpoints, identified previously14 in the same OE cells, to the OiRDs, according to transcription levels, as in a, with the analysis restricted to the early S replicating domains. Statistical comparisons, using random permutation analysis, are between observed (red) and genomic (grey) frequencies. NS, not significant. c, Mapping of genomic rearrangement (Rearr; n = 490,711) breakpoints from a TCGA pan-cancer data set4 to the OiRDs, according to transcription levels, as in a, with the analysis restricted to the early S replicating domains. Statistical comparisons, using z-scores, are between observed and genomic frequencies. d, Mapping of genomic rearrangement (Rearr; n = 490,711) breakpoints in common cancer types from a TCGA pan-cancer data set4 to the OiRDs, with the analysis restricted to the early S replicating domains. KIRC, kidney renal cell; COAD, colon adenocarcinoma; HNSC, head and neck squamous cell; UCEC, uterine cervix; GBM, glioblastoma multiformae; LUAD, lung adenocarcinoma; LUSC, lung squamous cell; BRCA, breast; BLCA, bladder; OV, ovary. Statistical comparisons, using z-scores, are between observed (red) and genomic (grey) frequencies. e, Distribution of cancer rearrangement breakpoints4 according to the replication timing data obtained from the REPLIseq experiment shown in Extended Data Fig. 2.
Extended Data Figure 10 Proposed mechanism for oncogene-induced DNA replication stress.
During the length of a normal G1 phase, transcription progressively inactivates intragenic origins, such that upon S phase entry origin firing is restricted to intergenic domains. Following oncogene activation, cells enter prematurely into S phase, before the inactivation of all intragenic origins. This results in bidirectional forks within highly transcribed genes, leading to conflicts between the replication and transcription machineries, fork collapse, DNA DSBs and genomic instability.
Supplementary information
Supplementary Figure 1
This file contains the gating strategy used in the flow cytometry analyses. (PDF 75 kb)
Supplementary Tables
This file contains Supplementary Tables 1-11. (XLSX 1084 kb)
Supplementary Data
This file contains all the scripts and datasets required to process and plot the replication and transcription sequencing data. (ZIP 9968 kb)
Rights and permissions
About this article
Cite this article
Macheret, M., Halazonetis, T. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116 (2018). https://doi.org/10.1038/nature25507
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25507
This article is cited by
-
Transcription–replication conflicts underlie sensitivity to PARP inhibitors
Nature (2024)
-
Oncogenic c-Myc induces replication stress by increasing cohesins chromatin occupancy in a CTCF-dependent manner
Nature Communications (2024)
-
Quantity and quality of minichromosome maintenance protein complexes couple replication licensing to genome integrity
Communications Biology (2024)
-
cGAS-STING pathway expression correlates with genomic instability and immune cell infiltration in breast cancer
npj Breast Cancer (2024)
-
RECQL4 is not critical for firing of human DNA replication origins
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.