Abstract
DNA methylation by de novo DNA methyltransferases 3A (DNMT3A) and 3B (DNMT3B) at cytosines is essential for genome regulation and development1,2. Dysregulation of this process is implicated in various diseases, notably cancer. However, the mechanisms underlying DNMT3 substrate recognition and enzymatic specificity remain elusive. Here we report a 2.65-ångström crystal structure of the DNMT3A–DNMT3L–DNA complex in which two DNMT3A monomers simultaneously attack two cytosine–phosphate–guanine (CpG) dinucleotides, with the target sites separated by 14 base pairs within the same DNA duplex. The DNMT3A–DNA interaction involves a target recognition domain, a catalytic loop, and DNMT3A homodimeric interface. Arg836 of the target recognition domain makes crucial contacts with CpG, ensuring DNMT3A enzymatic preference towards CpG sites in cells. Haematological cancer-associated somatic mutations of the substrate-binding residues decrease DNMT3A activity, induce CpG hypomethylation, and promote transformation of haematopoietic cells. Together, our study reveals the mechanistic basis for DNMT3A-mediated DNA methylation and establishes its aetiological link to human disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999)
Okano, M., Xie, S. & Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220 (1998)
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002)
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005)
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001)
Chedin, F., Lieber, M. R. & Hsieh, C. L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA 99, 16916–16921 (2002)
Hata, K., Okano, M., Lei, H. & Li, E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 (2002)
Robertson, K. D. DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 (2005)
Yang, L., Rau, R. & Goodell, M. A. DNMT3A in haematological malignancies. Nat. Rev. Cancer 15, 152–165 (2015)
Ley, T. J. et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 363, 2424–2433 (2010)
Guo, X. et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517, 640–644 (2015)
Jia, D., Jurkowska, R. Z., Zhang, X., Jeltsch, A. & Cheng, X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 (2007)
Jurkowska, R. Z. et al. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 36, 6656–6663 (2008)
Gowher, H. & Jeltsch, A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites. J. Mol. Biol. 309, 1201–1208 (2001)
Gowher, H. et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 357, 928–941 (2006)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Guo, J. U. et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 17, 215–222 (2014)
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009)
Chen, T., Tsujimoto, N. & Li, E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24, 9048–9058 (2004)
Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015)
Tatton-Brown, K. et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 46, 385–388 (2014)
Holz-Schietinger, C., Matje, D. M. & Reich, N. O. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J. Biol. Chem. 287, 30941–30951 (2012)
Kim, S. J. et al. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood 122, 4086–4089 (2013)
Lu, R. et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell 30, 92–107 (2016)
Russler-Germain, D. A. et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454 (2014)
Losman, J. A. et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 (2013)
Song, J., Rechkoblit, O., Bestor, T. H. & Patel, D. J. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 (2011)
Zhou, L. et al. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321, 591–599 (2002)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D . et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Yu, M. et al. A resource for cell line authentication, annotation and quality control. Nature 520, 307–311 (2015)
Wang, G. G. et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods 3, 287–293 (2006)
Volz, D. C. et al. Tris(1,3-dichloro-2-propyl)phosphate induces genome-wide hypomethylation within early zebrafish embryos. Environ. Sci. Technol. 50, 10255–10263 (2016)
Yu, Y. et al. Comprehensive assessment of oxidatively induced modifications of DNA in a rat model of human Wilson’s disease. Mol. Cell. Proteomics 15, 810–817 (2016)
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011)
Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369 (2014)
Acknowledgements
We thank X. Cheng for comments on the manuscript, M. Okano, J. Wang, and J.-A. Losman for providing reagents used in the study, and staff members at the Advanced Light Source, Lawrence Berkeley National Laboratory, and at the Advanced Photo Source, Argonne National Laboratory, for access to X-ray beamlines. We are also grateful for the support of University of North Carolina facilities including Genomics Core, which are partly supported by UNC Cancer Center Core Support Grant P30-CA016086. This work was supported by Kimmel Scholar Awards (to J.S. and G.G.W.), the March of Dimes Foundation (1-FY15-345 to J.S.), the DoD Peer-reviewed Cancer Research Program (W81XWH-14-1-0232 to G.G.W.), Gabrielle’s Angel Foundation for Cancer Research (to G.G.W.), Gilead Sciences Research Scholars Program in haematology/oncology (to G.G.W.), University Cancer Research Fund of the N.C. state (to G.G.W.), and the National Institutes of Health (1R35GM119721 to J.S.; R35GM124736 to S.B.R.; 5R21ES025392 to Y.W.; and 1R01CA215284, 1R01CA218600, and 1R01CA211336 to G.G.W.). G.G.W. is a Research Scholar of American Cancer Society and a Junior Faculty Scholar of American Society of Haematology. R.L. was supported by a Lymphoma Research Foundation postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
Z.-M.Z., R.L., P.W., Y.Y., D.C., L.G., S.L., D.J., and J.S. performed experiments. S.B.R. provided technical support. Y.W., G.G.W., and J.S. conceived and organized the study. Z.-M.Z., R.L., G.G.W., and J.S. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks A. Jeltsch, R. Xu and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Structures of the DNMT3A–DNMT3L tetramer in complex with the 10/11-mer DNA.
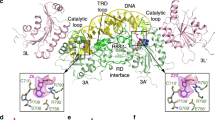
a, The sequence of the 10/11-mer DNA duplex used for structural study. b, Chemical formula of the covalent adduct of DNMT3A and 2′-deoxyzebularine. c, Data collection and refinement statistics. Each dataset was collected from a single crystal. d, Ribbon representations of the DNMT3A–DNMT3L tetramer in complex with the 10/11-mer DNA duplex and AdoHcy. DNMT3A, DNMT3L, and DNA are coloured in light blue, green, and wheat, respectively, and AdoHcy shown in sphere representation. The boxed areas show expanded views for the CpG sites (purple and yellow), the DNA-binding TRD and catalytic loops (left box), and the flipped out 2′-deoxyzebularine (dZ6′) surrounded by conserved catalytic residues (right box). The Fo − Fc omit map of 2′-deoxyzebularine (pink) is contoured at the 3σ level. The hydrogen-bonding interactions are depicted as dashed lines.
Extended Data Figure 2 Intermolecular interactions between the DNMT3A–DNMT3L tetramer and DNA.
a, Structural overlay of free DNMT3A–DNMT3L (PDB 2QRV) with the 10/11-mer DNA and 25-mer DNA-bound states. b, Stick representation of the 25-mer DNA duplex bound to the DNMT3A–DNMT3L tetramer, with the 2Fo − Fc omit map contoured at the 1σ level. c, The two helices of DNMT3A that interact with DNMT3L (shown in green) are coloured in pink (α4 and α5 in accordance with the numeration in Extended Data Fig. 3g) and preceded by two DNA contact loops, coloured in blue. The flipped out zebularine (Z5) is coloured in purple. The bound AdoHcy molecule is shown in sphere representation.
Extended Data Figure 3 Various intermolecular interactions between the two DNMT3A monomers and DNA.
a–f, DNA binding by the first and second DNMT3A monomer (defined as 3A and 3A′, respectively, in Fig. 1c) includes the intermolecular interactions between the TRD loop of DNMT3A and the DNA major groove (a, d), between the catalytic loop of DNMT3A and the DNA minor groove (b, e), and between the homodimeric interface of DNMT3A and the DNA backbone (c, f). The hydrogen-bonding interactions are shown as dashed lines. The water molecules are shown as purple spheres. g, Structure-based sequence alignment of DNMT3 proteins from human (hDNMT3A and hDNMT3B), mouse (mDnmt3a and mDnmt3b), and zebrafish (zDnmt3a and zDnmt3b). Completely conserved residues are coloured in white and highlighted in red. Partly conserved residues are coloured in red. Secondary structures are shown above the aligned sequences. The DNA-binding residues as revealed by this study are marked with black triangles.
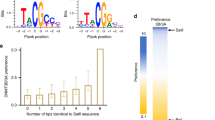
Extended Data Figure 4 The essential roles for the CpG-engaging residues of DNMT3A, R836, and V716 in DNMT3A-mediated CpG versus non-CpG methylations.
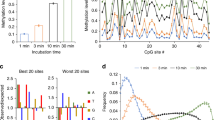
a, b, DNA methylation kinetics analysis of wild-type (DNMT3AWT, a) and R836A-mutated (DNMT3AR836A, b) DNMT3A using the CpG-, CpA-, or CpT-containing DNA substrates (n = 3 biological replicates). Purified DNMT3A–DNMT3L tetramer complex was used for measurements, followed by fitting with a first-order exponential equation. c, Ribbon representation of the crystal structure of DNMT3AR836A–DNMT3L tetramer in complex with the 25-mer DNA, with the CpG recognition by one of the DNMT3A monomers shown in expanded view. The 2Fo − Fc omit map of DNA was contoured at the 0.8σ level, and coloured in light blue. d, Methylation assay using either DNMT3AWT or DNMT3AV716G on CG-, CA-, and CT-containing DNA (n = 3 biological replicates). e, Immunoblots detect reconstituted expression of the indicated DNMT3A among TKO mouse ES cells, either the pooled stable-expression cell population or single-cell-derived clonal lines. EV, empty vector. A representative blot of two independent experiments is shown. For gel Source Data, see Supplementary Fig. 1. f, LC–MS analysis reveals the global 5-methyl-2′-deoxycytidine (5-mdC) levels (calculated as 5-mdC/2′-deoxyguanosine on the y axis) in the TKO ES cells after stable transduction of empty vector or the indicated DNMT3A (n = 3 biological replicates). Data are mean ± s.d.
Extended Data Figure 5 eRRBS reveals distribution of cytosine methylations in each sequence context among TKO ES cells with reconstituted expression of either DNMT3AWT or DNMT3AR836A.
a, The rates of bisulfite conversion for the indicated sequence context in each sample as determined by the unmethylated lambda DNA spike-in control. CpN, all cytosines. b, c, Pie charts showing the percentage of methylated cytosines (total number n shown at the bottom of each plot) identified among the DNMT3AWT- or DNMT3AR836A-expressing TKO ES cells in each sequence context. The methylated cytosines were called using a stringent binomial distribution-based filter to eliminate false positives from incomplete bisulfite conversion, with an FDR of 1% and 0.1% set for b and c, respectively. d, Distribution of methylation levels (percentage on x axis) for the indicated sequence context. Inserts show a closed view of the distribution at sites with intermediate to high levels of cytosine methylation.
Extended Data Figure 6 CpG and non-CpG methylations induced by DNMT3AWT versus DNMT3AR836A in the TKO ES cells.
a, Overall methylation levels of cytosines at four different sequence contexts as detected by eRRBS. On the y axis, the averaged methylation level of all cytosines within the mouse genome is shown, as calculated by normalization of the detected methylation cytosines over total cytosine numbers in the TKO ES cells reconstituted with either DNMT3AWT (left) or DNMT3AR836A (right). b, Global levels of CpG and CpH (H = A, C, or T) methylation induced by DNMT3AWT (green) or DNMT3AR836A (red) across the mouse chromosomes of the TKO ES cells. Box and whisker plots of 10-kb-bin-averaged methylation levels of each mouse chromosome are shown. c, Global levels of CpG and CpH methylation induced by DNMT3AWT (green) versus DNMT3AR836 (red) across all annotated genes. Each gene was divided into 100 equally sized bins and the 10-kb flanking region was divided into 50 equally sized bins. Averaged methylation levels were plotted for each bin. TSS, transcription start site; TES, transcription end site. d, Global levels of CpG and CpH methylation induced by DNMT3AWT (green) versus DNMT3AR836A (red) on the two opposite DNA strands. Boxplots of 10-kb-bin-averaged CpG, CpA, CpC, and CpT methylation levels of each strand are shown. e, Representative gene-wide views of CpG and CpH methylations at Foxp1 and Dock1, which are grouped into either the forward (+) or reverse (−) DNA strand. Cytosines covered by at least 15 reads from eRRBS data are shown, with each site designated by a vertical line. Panels a–e use the combined dataset of three biological replicates per group. Box plots depict the interquartile range, and whiskers depict 1.5 × interquartile range.
Extended Data Figure 7 Sanger bisulfite sequencing to validate the cytosine methylation levels mediated by DNMT3A, either wild type or defective in recognizing the CpG substrate, among the TKO mouse ES cells.
a, Sequence of the examined major satellite DNA region. Primers used for bisulfite PCR are denoted with 5′ and 3′ primer pairing. The counts for cytosines, highlighted in colour, are 9 for the CG dinucleotide, 33 for CA, 14 for CT, and 13 for CC. b, A representative result for bisulfite sequencing analysis of the major satellite repeat region described above in the TKO cells expressing empty vector, wild-type DNMT3A, or the indicated mutant. Each row represents one DNA clone and each column represents one site of cytosine, either methylated (filled) or unmethylated (open). c, d, Percentage of methylation mediated by DNMT3A or the indicated mutant at CpG (c) and non-CG (d) sites within the examined major satellite DNA region in the TKO ES cells. Data are mean ± s.d.; n = 4 independent bisulfite sequencing experiments as shown in b. e–h, Average cytosine methylation levels at each individual site grouped by the CpG (e), CpA (f), CpT (g), or CpC (h) context in the examined major satellite DNA among the TKO ES cells reconstituted with DNMT3AWT versus DNMT3AR836A (n = 4 biological replicates; mean ± s.d., with the labelled P values). Statistical analysis used a two-tailed Student’s t-test; NS, not significant.
Extended Data Figure 8 Effect of haematological cancer-associated mutations of DNMT3A on DNA methylation in vitro and in mouse TKO ES cells.
a, Methylation kinetics of DNMT3A with mutations located at the catalytic loop, compared with DNMT3AWT. b, Methylation kinetics of DNMT3A with an active site mutation, R792H. c, Methylation kinetics of DNMT3A with mutations located at the TRD loop. d, Methylation kinetics of DNMT3A with the hotspot mutation R882H. For a–d, DNMT3A–DNMT3L complex was used for the measurements (n = 3 biological replicates), followed by fitting with a first-order exponential equation. These data were measured independently from those shown in Fig. 4c. e, Immunoblot detects stable reconstitution of DNMT3AWT or the indicated DNMT3A mutant in the TKO ES cells. A representative blot of two independent experiments is shown. For gel Source Data, see Supplementary Fig. 1. f, LC–MS analysis reveals the global 5-methyl-2′- deoxycytidine (5-mdC) levels (calculated as 5-mdC/2′-deoxyguanosine) in the TKO ES cells after stable transduction of empty vector or the indicated DNMT3A. The methylation levels relative to TKO cells expressing DNMT3AWT are plotted. Data are mean ± s.d.; n = 4 biological replicates. g, Individual bisulfate sequencing detects the methylation level of cytosines in each sequence context within a major satellite DNA site at chromosome 2 in the TKO cells reconstituted with DNMT3AR836W (right), compared with DNMT3AWT (left) or DNMT3AR836A (middle; as determined in Fig. 3g) (n = 4; mean ± s.d.). Statistical analysis used a two-tailed Student’s t-test. h, In vitro methylation of CG-, CA-, or CT-containing DNA using DNMT3AWT (left) or DNMT3AR836W (right) in complex with DNMT3L, reacted for 40 min (n = 3 biological replicates). The methylation levels relative to CG-containing DNA substrates are plotted. Data are mean ± s.d.
Extended Data Figure 9 Effect of haematological cancer-associated mutations of DNMT3A on genomic DNA methylation in the TF-1 leukaemia cells.
a, Immunoblot of the TF-1 cells stably transduced with Myc-tagged DNMT3A, either wild type or the indicated cancer-associated mutants. EV, empty MSCV vector. A representative blot of two independent experiments is shown. For gel Source Data, see Supplementary Fig. 1. b, Profiling of the indicated DNMT3A-expressing TF-1 cell lines with the HumanMethylation_450K BeadChip array reveals the mean methylation β values for all examined CpGs. Each dot represents a biological replicate: that is, an independently derived stable-expression cell line (n = 3–8 biological replicates per group; mean ± s.d.). Statistical analysis used a two-tailed Student’s t-test. c, Density plot of methylation β values for all examined CpGs in the indicated DNMT3A-expressing TF-1 cell lines. The inserted box shows a zoom-in view for densities for highly methylated DNA sites among the indicated samples. Data are mean ± s.d., with the labelled P values. Statistical analysis used two-tailed Student’s t-test. d, Sanger bisulfite sequencing of the indicated regions from TF-1 cell lines stably transduced with empty vector, DNMT3AWT or the indicated cancer-associated mutant. Individual CpG sites (circles) are filled with black (methylated) or white (unmethylated). Red circles denote the CpG sites covered by the Illumina Infinium 450K DNA methylation array. Data are mean ± s.d.; n = 3 biological replicates. e, Methylation values of the indicated CpG sites (labelled by red circles in d) based on the measurements with the Infinium 450K DNA methylation arrays (n = 3–8 biological replicates; mean ± s.d.).
Extended Data Figure 10 Effect of haematological cancer-associated DNMT3A mutations on DNA hypomethylation and cytokine-independent growth of the TF-1 leukaemia cells.
a, Venn diagram of CpG sites with hypomethylation induced by either one of the three indicated strong DNA-binding-defective mutants of DNMT3A, V716D, K841E, and R792H. b, c, Bar plots (b) and scatter plots (c) showing methylation difference at the 605 commonly hypomethylated CpG sites identified in a among the TF-1 cells with stable transduction of either DNMT3AWT or the indicated mutant, compared with empty MSCV vector. A black line in b indicates empty vector control (n = 3–8 biological replicates per group; mean ± s.d.). In c, mean methylation β values are plotted of each individual CpG in the indicated DNMT3A experimental group (x axis) and control empty vector group (y axis; n = 3–8 biological replicates per group). d, e, Comparable occupancy of DNMT3A and its mutant forms at the indicated genomic loci with affected DNA methylation, as measured by ChIP analysis of the Myc-tagged DNMT3AWT or mutants in TF-1 stable-expression cell lines. Tested sites by ChIP–qPCR in d (n = 3 biological replicates; mean ± s.d.) included three CpG sites showing hypomethylation due to expression of mutant DNMT3A (left; also see Extended Data Fig. 9e), three sites showing hypermethylation due to expression of DNMT3AWT (middle; also see e, which shows mean methylation β values from measurements with the Infinium 450K DNA methylation arrays, n = 3–8 biological replicates; mean ± s.d.), and a negative control locus (right; the GAPDH transcription start site). The anti-Myc antibody was used for ChIP and the empty-vector-expressing TF-1 cells used as cell control for unspecific binding. f, Proliferation of the indicated DNMT3A stable-expression TF-1 cells in the presence of a supporting cytokine, GM-CSF (n = 2 biological replicates; mean ± s.d.). g, Proliferation of the indicated DNMT3A-expressing TF-1 cells after GM-CSF withdrawal (n = 3 biological replicates; mean ± s.d.).
Supplementary information
Supplementary Figure 1
This file contains the uncropped western blots from the main and supplementary figures. (PDF 169 kb)
Rights and permissions
About this article
Cite this article
Zhang, ZM., Lu, R., Wang, P. et al. Structural basis for DNMT3A-mediated de novo DNA methylation. Nature 554, 387–391 (2018). https://doi.org/10.1038/nature25477
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25477
This article is cited by
-
Intrafamily heterooligomerization as an emerging mechanism of methyltransferase regulation
Epigenetics & Chromatin (2024)
-
Structure-guided functional suppression of AML-associated DNMT3A hotspot mutations
Nature Communications (2024)
-
DNA methylation regulator-mediated modification patterns and risk of intracranial aneurysm: a multi-omics and epigenome-wide association study integrating machine learning, Mendelian randomization, eQTL and mQTL data
Journal of Translational Medicine (2023)
-
5mC modification orchestrates choriogenesis and fertilization by preventing prolonged ftz-f1 expression
Nature Communications (2023)
-
Base editor scanning charts the DNMT3A activity landscape
Nature Chemical Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.