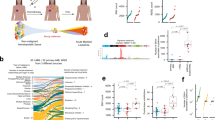
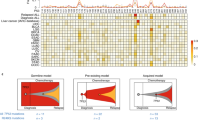
Abstract
Relapsed acute lymphoblastic leukaemia (ALL) is associated with resistance to chemotherapy and poor prognosis1. Gain-of-function mutations in the 5′-nucleotidase, cytosolic II (NT5C2) gene induce resistance to 6-mercaptopurine and are selectively present in relapsed ALL2,3. Yet, the mechanisms involved in NT5C2 mutation-driven clonal evolution during the initiation of leukaemia, disease progression and relapse remain unknown. Here we use a conditional-and-inducible leukaemia model to demonstrate that expression of NT5C2(R367Q), a highly prevalent relapsed-ALL NT5C2 mutation, induces resistance to chemotherapy with 6-mercaptopurine at the cost of impaired leukaemia cell growth and leukaemia-initiating cell activity. The loss-of-fitness phenotype of NT5C2+/R367Q mutant cells is associated with excess export of purines to the extracellular space and depletion of the intracellular purine-nucleotide pool. Consequently, blocking guanosine synthesis by inhibition of inosine-5′-monophosphate dehydrogenase (IMPDH) induced increased cytotoxicity against NT5C2-mutant leukaemia lymphoblasts. These results identify the fitness cost of NT5C2 mutation and resistance to chemotherapy as key evolutionary drivers that shape clonal evolution in relapsed ALL and support a role for IMPDH inhibition in the treatment of ALL.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hunger, S. P. & Mullighan, C. G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 373, 1541–1552 (2015)
Tzoneva, G. et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 19, 368–371 (2013)
Meyer, J. A. et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat. Genet. 45, 290–294 (2013)
Gerby, B. et al. Expression of CD34 and CD7 on human T-cell acute lymphoblastic leukemia discriminates functionally heterogeneous cell populations. Leukemia 25, 1249–1258 (2011)
Cox, C. V., Diamanti, P., Evely, R. S., Kearns, P. R. & Blair, A. Expression of CD133 on leukemia-initiating cells in childhood ALL. Blood 113, 3287–3296 (2009)
Konopleva, M. et al. Stromal cells prevent apoptosis of AML cells by up-regulation of anti-apoptotic proteins. Leukemia 16, 1713–1724 (2002)
Hawkins, E. D. et al. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538, 518–522 (2016)
Mullighan, C. G. et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380 (2008)
Ma, X. et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat. Commun. 6, 6604 (2015)
Oshima, K. et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc. Natl Acad. Sci. USA 113, 11306–11311 (2016)
Li, B. et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat. Med. 21, 563–571 (2015)
Mullighan, C. G. et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471, 235–239 (2011)
Malinowska-Ozdowy, K. et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia 29, 1656–1667 (2015)
Spychała, J., Madrid-Marina, V. & Fox, I. H. High Km soluble 5′-nucleotidase from human placenta. Properties and allosteric regulation by IMP and ATP. J. Biol. Chem. 263, 18759–18765 (1988)
Gazziola, C., Ferraro, P., Moras, M., Reichard, P. & Bianchi, V. Cytosolic high Km 5′-nucleotidase and 5′(3′)-deoxyribonucleotidase in substrate cycles involved in nucleotide metabolism. J. Biol. Chem. 276, 6185–6190 (2001)
Brouwer, C. et al. Role of 5′-nucleotidase in thiopurine metabolism: enzyme kinetic profile and association with thio-GMP levels in patients with acute lymphoblastic leukemia during 6-mercaptopurine treatment. Clin. Chim. Acta 361, 95–103 (2005)
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998)
Herranz, D. et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 20, 1130–1137 (2014)
Ferrando, A. A. & López-Otín, C. Clonal evolution in leukemia. Nat. Med. 23, 1135–1145 (2017)
Clappier, E. et al. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 208, 653–661 (2011)
Wong, T. N. et al. Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood 127, 893–897 (2016)
Shlush, L. I. et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017)
Andersson, D. I. & Hughes, D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010)
Greaves, M. & Maley, C. C. Clonal evolution in cancer. Nature 481, 306–313 (2012)
Wang, J. et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. eLife 3, (2014)
Gan, L. et al. The immunosuppressive agent mizoribine monophosphate forms a transition state analogue complex with inosine monophosphate dehydrogenase. Biochemistry 42, 857–863 (2003)
Reaves, M. L., Young, B. D., Hosios, A. M., Xu, Y. F. & Rabinowitz, J. D. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature 500, 237–241 (2013)
Ser, Z. et al. Targeting one carbon metabolism with an antimetabolite disrupts pyrimidine homeostasis and induces nucleotide overflow. Cell Reports 15, 2367–2376 (2016)
Palomero, T. et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia 20, 1279–1287 (2006)
Kimbrel, E. A . et al. Systematic in vivo structure-function analysis of p300 in hematopoiesis. Blood 114, 4804–4812 (2009)
Guo, K. et al. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology 148, 3987–3997 (2007)
Herranz, D. et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat. Med. 21, 1182–1189 (2015)
Iacobucci, I. et al. Truncating erythropoietin receptor rearrangements in acute lymphoblastic leukemia. Cancer Cell 29, 186–200 (2016)
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012)
Kennedy, S. R. et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc. 9, 2586–2606 (2014)
Futreal, P. A. et al. A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004)
Van Vlierberghe, P. & Ferrando, A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Invest. 122, 3398–3406 (2012)
Zhang, J. et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Blood 118, 3080–3087 (2011)
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009)
Acknowledgements
We are grateful to R. Kopan for the ΔE-NOTCH1 construct and T. Ludwig for the ROSA26Cre-ERT2/+ mouse. This work was supported by the Leukemia & Lymphoma Society Quest for Cures (R0749-14) and Translational Research (6455-15; 6531-18) awards (A.A.F.), an Innovative Research Award from the Alex Lemonade Stand Foundation (A.A.F.), the Chemotherapy Foundation (A.A.F.), National Institutes of Health (NIH) grants R35 CA210065 (A.A.F.), R01 CA206501 (A.A.F.), U54 CA193313 (R.R.), R01 CA185486 (R.R.), U54 CA209997 (R.R.), U10 CA98543 (J.M.G., M.L.L.), P30 CA013696, the Human Specimen Banking Grant U24 CA114766 (J.M.G.), the Stewart Foundation (R.R.) and the American Lebanese Syrian Associated Charities of St Jude Children’s Research Hospital. G.T. was supported by a HHMI International Student Research Fellowship. M.S.M. was supported by a Rally Foundation fellowship. C.L.D. was supported by NIH/NCI T32-CA09503. J.Y. was supported by the China Scholarship Council (CSC 201304910347) and the Ter Meulen Grant of the Royal Netherlands Academy of Arts and Sciences. E.W. was supported by the Dutch Cancer Society (KUN2012-5366).
Author information
Authors and Affiliations
Contributions
G.T. and C.L.D. performed biochemical, cellular and animal studies. M.S.-M. and K.O. helped in experimental therapeutic experiments. A.A.-I. and H.K. analysed deep sequencing data. C.J.M. performed ISN analysis. M.L.S., M.K., K.K., M.P., G.B., J.M.G.-F. and M.L.L. provided clinical specimens. J.Y., E.W. and I.I. performed and analysed droplet PCR analyses. R.K.-S. provided clinical samples and correlative analyses of clinical data. C.G.M. supervised droplet PCR analyses; R.R. supervised deep sequencing and ISN analyses. A.A.F. designed the study, supervised the research and wrote the manuscript with G.T. and C.L.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Schematic representation of 6-MP activation and mechanism of action.
The hypoxanthine-guanine phosphoribosyl transferase enzyme (HPRT) processes 6-MP to thio-IMP, which is then converted to thio-XMP and thio-GMP. Subsequent metabolism of thio-GMP by kinases and reductases yields thio-dGTP which is incorporated into replicating DNA strands and triggers the DNA mismatch-repair machinery, leading to cell cycle arrest and apoptosis. The anti-leukaemic effects of 6-MP are in part also attributed to a second metabolic pathway in which thiopurine S-methyl transferase (TPMT) methylates thio-IMP to form methylthio-IMP (MeTIMP), which is a potent inhibitor of amidophosphoribosyltransferase (ATase), an enzyme catalysing the committed step of de novo purine biosynthesis.
Extended Data Figure 2 Conditional knock-in targeting of Nt5c2, generation and analysis of a Nt5c2(R367Q) conditional inducible T-ALL model.
a, Schematic representation of the targeting strategy for the generation of Nt5c2+/co-R367Q conditional knock-in mice. b, Southern blot analysis of DNA samples from Nt5c2+/+ and targeted Nt5c2+/co-R367Q embryonic stem cells after digestions with BamHI restriction enzyme and hybridization of a DNA probe external to the long arm. c, Southern blot analysis of DNA samples from Nt5c2+/+ and targeted Nt5c2+/co-R367Q embryonic stem cells after digestion with ApaI restriction enzyme and hybridization of a DNA probe to the short arm. d, Schematic depiction of the strategy for developing conditional inducible Nt5c2+/co-R367Q primary mouse T-ALL tumours and for assessing the role of Nt5c2+/R367Q on leukaemia progression and response to chemotherapy. e, Representative FACS plot of a Rosa26+/creERT2Nt5c2+/co-R367Q ΔE-NOTCH1-induced primary T-ALL tumour with a CD4+CD8+ immunophenotype. f, Representative genotyping PCR results from genomic DNA of a Rosa26+/CreERT2 Nt5c2+/co-R367Q ΔE-NOTCH1-induced primary T-ALL tumour treated with 4-hydroxytamoxifen (TMX) or vehicle only (ethanol, ETOH) in vitro showing Cre-mediated deletion of the exon 14–18 Nt5c2 wild-type mini-gene. g, Tumour burden assessed in the spleen (percentage of GFP+ cells) in mice allografted with NOTCH1-induced Nt5c2+/co-R367Q and isogenic Nt5c2+/R367Q primary leukaemia cells treated with a range of 6-MP doses (n = 5 per group). h, Analysis of selection for the mutant allele encoding Nt5c2(R367Q) by qPCR in mice allografted with Nt5c2+/co-R367Q and Nt5c2+/R367Q primary mouse T-ALL cells at a 1:10, 1:100 and 1:1,000 Nt5c2+/R367Q:Nt5c2+/co-R367Q dilution and treated with vehicle or 6-MP (n = 5 mice per group and n = 3 technical replicates for the controls). The horizontal bar represents mean values. P values were calculated using two-tailed Student’s t-test (g) or a one-tailed Student’s t-test (h). Data in e and f show representative results from more than two experiments.
Extended Data Figure 3 Decreased expression of the allele encoding Nt5c2(R367Q) allele upon leukaemia progression in vivo.
Sanger sequencing chromatograms of cDNA from tumours in Fig. 2c show decreased expression of the Nt5c2(R367Q)-encoding allele over the wild-type Nt5c2 allele compared with recently 4-hydroxytamoxifen treated Rosa26+/creERT2Nt5c2+/co-R367Q cells (Fig. 1a). Mutant-allele deoxynucleotides are indicated in red.
Extended Data Figure 4 NT5C2(R367Q) expression leads to increased purine export in T-ALL and B-ALL cell lines.
Diagram of the purine de novo biosynthesis and salvage pathways, showing gas chromatography–mass spectrometry and liquid chromatography–tandem mass spectrometry metabolic profiles (mass spectrometry scaled intensity, arbitrary units) of CUTLL1 and REH cell lines expressing wild-type NT5C2 or NT5C2(R367Q) and their corresponding conditioned media (n = 3 biological replicates per sample). Box plots represent the upper quartile to lower quartile distribution. Plus signs indicate mean values, horizontal lines indicate median values and whiskers indicate the maximum and minimum values of the distributions.
Extended Data Figure 5 NT5C2 mutations are late events in ALL.
ISN illustrating the sequential order of somatic mutations in relapsed ALL by pooling evolutionary paths across patients. Each node represents a gene and each arrow points from a gene with an early event to a gene with a late event. To test whether a gene within the ISN was significantly early or late, we used a one-sided binomial test based on the in-degree and out-degree of each node.
Extended Data Figure 6 Guanosine rescue of mizoribine sensitivity in vitro and mizoribine activity against NT5C2(R367Q) mutant cells in vivo.
a, b, Cell viability assays showing drug responses of wild-type Nt5c2+/co-R367Q primary mouse T-ALL cells (a) or mutant Nt5c2+/R367Q mouse T-ALL lymphoblasts (b) to increasing doses of mizoribine in the presence of 20 μM guanosine (n = 3 biological replicates). c, Analysis of tumour burden assessed by bioimaging in mice transplanted with wild-type Nt5c2+/co-R367Q leukaemia cells (left flank) or mutant Nt5c2+/R367Q leukaemia cells (right flank) treated with a range of mizoribine doses (n = 8 mice for the vehicle group and n = 4 mice per treated group). d, Quantification of data from c. e, Analysis of tumour burden assessed by spleen weight in mice allografted with wild-type NT5C2 ALL-4 diagnosis or NT5C2(R367Q) ALL-4 relapsed-patient derived leukaemia cells treated with 100 mg kg−1 mizoribine twice a day (n = 6 for diagnosis vehicle group, n = 3 for relapse treated group and n = 7 for diagnosis treated and relapse vehicle groups). f, Analysis of tumour burden assessed by percentage of CD45+ cells in the bone marrow of mice allografted with NT5C2 wild-type ALL-4 diagnosis or NT5C2(R367Q) ALL-4 relapsed-patient derived leukaemia cells treated with 100 mg kg-1 mizoribine twice daily. (n = 3–7 mice per group). Horizontal bars in a, b, d–f indicate mean values. P values were calculated using a two-tailed Student’s t-test.
Extended Data Figure 7 6-MP and IMPDH inhibition response in CUTLL1 cells.
a, Cell viability assays showing drug responses of the CUTLL1 cell line infected with wild-type or mutant NT5C2-expressing lentiviruses to increasing doses of 6-MP. b, Cell viability assays as in a documenting the response to mizoribine. c, d, Cell viability assay showing drug responses of wild-type (c) or NT5C2(R367Q) (d) CUTLL1 cells to increasing doses of mizoribine in the presence of 20 μM guanosine. e, Growth curve of CUTLL1 cells infected with a control short hairpin RNA (shRNA) targeting GFP or an shRNA targeting IMPDH2. f, Growth curve of wild-type or NT5C2(R367Q) CUTLL1 cells and infected with a shRNA targeting IMPDH2. a–f, Data are from three biological replicates. Horizontal bars in c and d indicate mean values. P values were calculated using a two-tailed Student’s t-test. *P ≤ 0.05.
Extended Data Figure 8 6-MP and IMPDH inhibition response in REH B-ALL cells.
a, Cell viability assay showing drug responses of the REH cell line infected with wild-type or mutant NT5C2-expressing lentiviruses to increasing doses of 6-MP. b, Cell viability assays as in a documenting the response to mizoribine. c, d, Cell viability assay showing drug responses of wild-type (c) or NT5C2(R367Q) (d) REH cells to increasing doses of mizoribine in the presence of 20 μM guanosine. e, Growth curve of REH cells infected with a control shRNA targeting GFP or shRNA targeting IMPDH2. f, Growth curve of wild-type or NT5C2(R367Q) REH cells and infected with an shRNA targeting IMPDH2. a–f, n = 3 biological replicates. Horizontal bars in c and d indicate mean values. P values were calculated using a two-tailed Student’s t-test. *P ≤ 0.05.
Supplementary information
Supplementary Table 1
Metabolomic Analysis of Nt5c2+/R367Q and Nt5c2+/co-R367Q ALL lymphoblasts. (XLSX 187 kb)
Supplementary Table 2
Metabolomic Analysis of Nt5c2+/R367Q and Nt5c2+/co-R367Q ALL conditioned media. (XLSX 111 kb)
Supplementary Table 3
Metabolomic Analysis of NT5C2 WT and NT5C2 R367Q expressing CUTLL1 and REH Cells. (XLSX 447 kb)
Supplementary Table 4
Metabolomic Analysis of Conditioned Media from NT5C2 WT and NT5C2 R367Q expressing CUTLL1 and REH Cells. (XLSX 199 kb)
Rights and permissions
About this article
Cite this article
Tzoneva, G., Dieck, C., Oshima, K. et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature 553, 511–514 (2018). https://doi.org/10.1038/nature25186
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25186
This article is cited by
-
Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma
npj Precision Oncology (2024)
-
Targetable lesions and proteomes predict therapy sensitivity through disease evolution in pediatric acute lymphoblastic leukemia
Nature Communications (2023)
-
In vivo PDX CRISPR/Cas9 screens reveal mutual therapeutic targets to overcome heterogeneous acquired chemo-resistance
Leukemia (2022)
-
Maintenance therapy for acute lymphoblastic leukemia: basic science and clinical translations
Leukemia (2022)
-
Phosphoproteomic profiling of T cell acute lymphoblastic leukemia reveals targetable kinases and combination treatment strategies
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.