Abstract
Inhibitors of Mek1/2 and Gsk3β, known as 2i, enhance the derivation of embryonic stem (ES) cells and promote ground-state pluripotency in rodents1,2. Here we show that the derivation of female mouse ES cells in the presence of 2i and leukaemia inhibitory factor (2i/L ES cells) results in a widespread loss of DNA methylation, including a massive erasure of genomic imprints. Despite this global loss of DNA methylation, early-passage 2i/L ES cells efficiently differentiate into somatic cells, and this process requires genome-wide de novo DNA methylation. However, the majority of imprinting control regions (ICRs) remain unmethylated in 2i/L-ES-cell-derived differentiated cells. Consistently, 2i/L ES cells exhibit impaired autonomous embryonic and placental development by tetraploid embryo complementation or nuclear transplantation. We identified the derivation conditions of female ES cells that display 2i/L-ES-cell-like transcriptional signatures while preserving gamete-derived DNA methylation and autonomous developmental potential. Upon prolonged culture, however, female ES cells exhibited ICR demethylation regardless of culture conditions. Our results provide insights into the derivation of female ES cells reminiscent of the inner cell mass of preimplantation embryos.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nichols, J. & Smith, A. Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 (2009)
Hackett, J. A. & Surani, M. A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell 15, 416–430 (2014)
Smith, Z. D. et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 (2012)
Ying, Q. L. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008)
Ficz, G. et al. FGF signaling inhibition in ES cells drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359 (2013)
Habibi, E. et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360–369 (2013)
Hackett, J. A. et al. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Reports 1, 518–531 (2013)
Leitch, H. G. et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311–316 (2013)
Marks, H. et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590–604 (2012)
Buehr, M. et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 (2008)
Czechanski, A. et al. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protocols 9, 559–574 (2014)
Guo, G. et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports 6, 437–446 (2016)
Takada, T. et al. The ancestor of extant Japanese fancy mice contributed to the mosaic genomes of classical inbred strains. Genome Res. 23, 1329–1338 (2013)
Zvetkova, I. et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat. Genet. 37, 1274–1279 (2005)
Choi, J. et al. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell 20, 706–719.e7 (2017)
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992)
Holm, T. M. et al. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8, 275–285 (2005)
Tucker, K. L. et al. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10, 1008–1020 (1996)
Theunissen, T. W. et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19, 502–515 (2016)
Kato, Y. et al. Developmental potential of mouse primordial germ cells. Development 126, 1823–1832 (1999)
Ohnishi, K. et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156, 663–677 (2014)
Stadtfeld, M. et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 (2010)
Wakayama, T., Perry, A. C., Zuccotti, M., Johnson, K. R. & Yanagimachi, R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374 (1998)
Branco, M. R. et al. Maternal DNA methylation regulates early trophoblast development. Dev. Cell 36, 152–163 (2016)
Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl Acad. Sci. USA 90, 8424–8428 (1993)
Niwa, H. et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 (2005)
Schulz, E. G. et al. The two active X chromosomes in female ES cells block exit from the pluripotent state by modulating the ES cell signaling network. Cell Stem Cell 14, 203–216 (2014)
Shirane, K. et al. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell 39, 87–103 (2016)
Shimizu, T. et al. Dual inhibition of Src and GSK3 maintains mouse embryonic stem cells, whose differentiation is mechanically regulated by Src signaling. Stem Cells 30, 1394–1404 (2012)
Pastor, W. A. et al. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18, 323–329 (2016)
Takada, T., Yoshiki, A., Obata, Y., Yamazaki, Y. & Shiroishi, T. NIG_MoG: a mouse genome navigator for exploring intersubspecific genetic polymorphisms. Mamm. Genome 26, 331–337 (2015)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Kim, S. I. et al. Inducible transgene expression in human iPS cells using versatile all-in-one piggyBac transposons. Methods Mol. Biol. 1357, 111–131 (2016)
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011)
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012)
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011)
Rosenbloom, K. R. et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681 (2015)
Illingworth, R. S. et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6, e1001134 (2010)
Kobayashi, H. et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440 (2012)
Tomizawa, S. et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 138, 811–820 (2011)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010)
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013)
Wakayama, T., Rodriguez, I., Perry, A. C., Yanagimachi, R. & Mombaerts, P. Mice cloned from embryonic stem cells. Proc. Natl Acad. Sci. USA 96, 14984–14989 (1999)
Terashita, Y. et al. Latrunculin A can improve the birth rate of cloned mice and simplify the nuclear transfer protocol by gently inhibiting actin polymerization. Biol. Reprod. 86, 180 (2012)
Terashita, Y. et al. Latrunculin A treatment prevents abnormal chromosome segregation for successful development of cloned embryos. PLoS One 8, e78380 (2013)
Acknowledgements
We are grateful to T. Shiroishi, M. Saitou and S. Yokobayashi for helpful suggestions, K. Woltjen for providing piggyBac vectors, P. Karagiannis for critical reading of this manuscript, and T. Ukai, M. Kabata, S. Sakurai, D. Seki and T. Sato for technical assistance. Y.Y. was supported in part by P-CREATE, SICORP, Japan Agency for Medical Research and Development (AMED); JSPS KAKENHI 15H04721; the Princess Takamatsu Cancer Research Fund; the Takeda Science Foundation; and the Naito Foundation. Y.Y. and T.Y. were supported by Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine, AMED. T.Y. was supported by AMED-CREST; JSPS KAKENHI 15H01352; and iPS Cell Research Fund. M.Y. was supported by JSPS KAKENHI 15J05792.
Author information
Authors and Affiliations
Contributions
M.Y. and Y.Y. designed and conceived the study and wrote the manuscript. M.Y. generated cell lines, performed experiments, analysed microarray data and generated WGBS and methyl-seq libraries. S.K., E.M., S.W. and T.W. performed nuclear transfer. K.S. provided technical instructions. A.T., S.K., S.W. and T.W. performed 2n and 4n blastocyst injections. T.Y. analysed all the WGBS, methyl-seq and RNA-seq data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Zwaka and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 WGBS for evaluation of global and ICR methylation.
The number of sequencing reads required to analyse global and ICR methylations. Sequenced reads acquired by NextSeq at single read were used for the evaluation. The indicated number of sequenced reads were randomly selected, followed by determination of global and ICR methylations. The determination was repeated 100 times. a, Mean ± s.d. of the 100 independent data for global DNA methylation are shown. The methylation data using sequenced reads by HighSeq (>100M reads for each sample) are shown as a control. b, A box-and-whisker plot of the 100 independent data for ICR methylation. Solid lines in each box indicate the median. Bottom and top of the box are lower and upper quartiles, respectively. Whiskers extend to ±1.5 interquartile range (IQR). c, The number of sequence reads for all samples used in this study.
Extended Data Figure 2 Characterization of 2i/L ES cells and S/L ES cells.
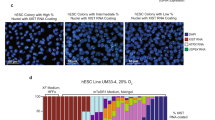
a, Global CHG and CHH methylation percentages were measured by WGBS in 2i/L ES cells and S/L ES cells. DNA methylation levels are lower in 2i/L ES cells than in S/L ES cells. Two independent clones for XX and XY were analysed for 2i/L ES cells and S/L ES cells. b, Immunostaining for 5mC in 2i/L ES cells (#2, XX) and S/L ES cells (#8, XX). Staining signals are hardly detectable in 2i/L ES cells. Scale bars, 200 μm. c, Karyotype analysis by Q-band in 2i/L ES cells (#1 and #2, XX, p3) and S/L ES cells (#8 and #9, XX, p3) shows 40 chromosomes including two X chromosomes (20 cells were analysed for karyotype analysis). d, Dynamics of whole genome and ICR methylation levels during female 2i/L ES cell derivation (p1, p2, p3). F1 (129/C57BL6) blastocysts were used for derivation of 2i/L ES cells. Note that a substantial reduction in both global and ICR methylation levels is already detectable at p1. The methylation levels are further reduced after passage. Independent three XX 2i/L ES cell clones were analysed. Conventional bisulfite sequencing at Nespas-Gnasxl and Snrpn ICR confirmed reduced ICR methylation at p1. Parental alleles were distinguished by SNPs (129 and C57BL6). e, Genome browser tracks showing representative loci that exhibit decreased methylation in 2i/L ES cells. DNA methylation in 2i/L ES cells (#15, XX, #23, XY) and S/L ES cells (#36, XX) are shown. Location of RefSeq genes and gene symbols for representative genes are indicated below. f, CpG methylation at U2af1-rs1 (also known as Zrsr1) of 2i/L ES cells (#15, XX, #23, XY), S/L ES cells (#36, XX) and MEFs (XX). Both 2i/L ES cells and S/L ES cells revealed a substantial reduction of ICR methylation. Each black dot represents a methylation percentage for each CpG site. Red and blue dots indicate methylation levels at the maternal 129 allele and paternal MSM allele, respectively.
Extended Data Figure 3 DNA methylation analyses of 2i/L ES cells, S/L ES cells and MEFs.
a, Conventional bisulfite sequencing at Peg10 DMR in multiple clones. Parental alleles were distinguished by SNPs. Open circles represent unmethylated CpGs, and closed circles represent methylated CpGs. Massive loss of CpG methylation is detected in female 2i/L ES cell lines (#1, #2, #12 and #15, XX), and erosion is observed in male 2i/L ES cells (#23, XY), whereas S/L ES cell lines (#9, #36, XX and #12, XY) often retain DMR methylation. b, Heat map for ICR methylation in male ES cells. ICR methylation status in 2i/L ES cells (#2, XX), 2i/L ES cells (#23, XY) and control MEFs (XY) are shown. The heat map depicts the methylation status at CpG sites in which parental alleles have been distinguished. CpG methylation levels and allelic balance for the methylation are shown for each CpG site. Both male and female 2i/L ES cells (#23, XY and #2, XX) exhibit decreased DNA methylation at ICRs. Colour scale is shown for DNA methylation levels and allelic balance. c, Sex differences in ICR methylation in MEFs and 2i/L ES cells. Methylation percentages of each CpG site are shown in a heat map. CpG methylation was measured by methyl-seq. Preferential reduction of ICR methylation in female cells is not observed in MEFs, while female 2i/L ES cells exhibit prominent ICR demethylation. Colour scale shows frequencies of the methylation value at each CpG site. d, Allelic expression analysis of the Peg10 gene in MEFs (XY), 2i/L ES cells (#2, XX) and S/L ES cells (#36, XX). 2i/L ES cells exhibit biallelic expression of Peg10. Numbers of subcloned maternal/paternal alleles in RT–PCR are shown. e, 2i/L ES cells (#15, XX) are capable of forming teratomas, which include three different germ cell lineages (scale bars, 50 μm). f, GFP-labelled 2i/L ES cells (#15, XX) efficiently contributed to E14.5 embryos (scale bars of ES cells, 100 μm; scale bars of embryo, 10 mm). GFP-positive MEFs were derived from one chimaeric embryo by sorting and used for the DNA methylation analysis of 2i/L-MEFs. g, Genome browser tracks showing representative loci that exhibit widespread de novo methylation in 2i/L-S/L ES cells and 2i/L-MEFs. DNA methylation in 2i/L ES cells (#15, XX), 2i/L-S/L ES cells (#15, XX), 2i/L-MEFs (#15, XX), S/L ES cells (#36, XX) and control MEFs (XX) are shown. Location of RefSeq genes and gene symbols for representative genes are indicated below. Methyl-seq data were used for the analysis.
Extended Data Figure 4 Targeted disruption of Dnmt genes in 2i/L ES cells.
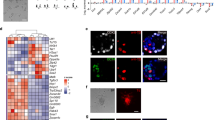
a, Strategy of knockout for DNA methyltransferases using CRISPR–Cas9. Each gRNA targeting sequence is underlined, and the PAM sequence is labelled in green. 2i/L ES cells (#15, XX) were transfected with plasmids expressing CRISPR–Cas9 and gRNA in 2i/L culture condition. Dnmt3a was disrupted in Dnmt3b−/− ES cells to obtain double-knockout ES cells for Dnmt3a and Dnmt3b. b, Sanger sequencing revealed frameshift mutations in both alleles for all knockout cells. c, Western blotting analysis for Dnmt3a, Dnmt3b, Dnmt3l and Dnmt1 in 2i/L ES cells (wild type, #15, XX), Dnmt triple-knockout ES cells in serum32, Dnmt3a−/−, Dnmt3b−/−, Dnmt3l−/−, Dnmt1−/− and Dnmt3a/3b−/− 2i/L ES cells (#15, XX). For the detection of Dnmt3a and Dnmt3b, ES cells were cultivated in DMEM/F12 and Neurobasal supplemented with B27, N2 cell-supplements, 20 ng ml−1 recombinant human activin A (R&D Systems), 12 ng ml−1 bFGF (wako), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 1% knockout serum replacement (GIBCO) for 2 days. d, Growth assay of Dnmt-knockout 2i/L ES cell lines cultured in the absence of 2i/L medium. Cell numbers were counted at day 0, day 2, day 4 and day 6 (independent experiments, n = 3). Data are presented as mean ± s.d. Dnmt1−/− and Dnmt3a/3b−/− 2i/L ES cells stopped their growth, while individual Dnmt3-knockout 2i/L ES cells had normal growth. e, Chimaeric contribution of Dnmt-knockout 2i/L ES cells in E14.5 embryos. For each ES cell line (p5–7), 7–8 GFP-labelled cells were injected into a 2n blastocyst, and 20 embryos were transferred into a pseudo-pregnant mouse. The percentage of chimaeras was calculated from the number of GFP-positive embryos out of total embryos. Scale bars, 10 mm.
Extended Data Figure 5 Allelic analysis of DMR methylation and gene expression.
a, Individual CpG methylation levels are shown as black dots for ICRs at Zac1 and Grb10. Red and blue dots indicate methylation levels at the maternal 129 allele and paternal MSM allele, respectively. Note that ICRs remain unmethylated in 2i/L-S/L ES cells (#15, XX) and 2i/L-MEFs (#15, XX). b, CpG methylation percentage at ICRs measured by WGBS in 2i/L-MEFs and S/L-MEFs. S/L-MEFs were derived from GFP-labelled S/L ES cells (#36, XX) at p5. Note that 2i/L-MEFs do not gain de novo methylation at ICRs, whereas S/L-MEFs maintain ICR methylation. c, Allelic expression analysis of the Peg10 gene in 2i/L-S/L ES cells (#15, XX) and 2i/L-MEFs (#15, XX). Biallelic expression of Peg10 is detectable in both 2i/L-S/L ES cells and 2i/L-MEFs. Numbers of subcloned maternal/paternal alleles in RT–PCR are shown. d, Heat map for methylation status at CpG sites in oocyte/sperm DMRs40. CpG methylation levels and allelic balance in 2i/L ES cells (#15, XX), 2i/L-S/L ES cells (#15, XX), 2i/L-MEFs (#15, XX), S/L ES cells (#36, XX) and control MEFs (XX) are shown. CpG sites in which parental alleles have been distinguished are included in the heat map. Most CpG sites in oocyte/sperm DMRs, except for ICRs, show de novo methylation. e, Aberrant de novo methylation at secondary DMR in 2i/L-MEFs. The genomic location of ICR (black) and secondary DMR (green) are shown below. f, Slight increase in CpG methylation is observed in a subset of ICRs. The increased methylation lacks allelic specificity.
Extended Data Figure 6 Comparison of developmental potential of 2i/L ES cells and S/L ES cells.
a, Efficient contribution of 2i/L ES cells to E14.5 chimaeric embryos. GFP-labelled 2i/L ES cells (#1, XX and #2, XX, p5–6) and S/L ES cells (#8, XX and #9, XX, p5–6) were injected into diploid blastocysts, and the injected embryos were transplanted into the uteri of pseudo-pregnant mice. The number of obtained embryos for 2i/L ES cells #1, 2i/L ES cells #2, S/L ES cells #8 and S/L ES cells #9 is 5, 10, 12 and 8, respectively. Scale bars, 10 mm. b, Table shows results of nuclear transfer, including the number of cells that formed pronuclei (PN), developed into 2-cell-stage embryos, used for embryo transfer (ET) and produced cloned pups. c, E10.5 cloned embryo from 2i/L ES cell nucleus shows growth retardation phenotype. d, Representative image of cloned pups derived from S/L ES cells. Cloned pups were obtained only from S/L ES cells (#8, XX and #9, XX). e, Histological analysis of the cloned placentae (E10.5). Representative histological images of the cloned placentae are shown. Scale bars, 500 μm. Hypoplasia of the labyrinth layer and focal necrosis (arrows) were detectable in the placentae from 2i/L ES cell nuclei. Number of cloned placentae examined: 2i/L ES cells #1; n = 8, 2i/L ES cells #2; n = 11, S/L ES cells #8; n = 7, and S/L ES cells #9; n = 9. f, Immunostaining reveals the reduced expression of E-cadherin in 2i/L-ES-cell-cloned placentae. Hypoplasia of the labyrinth layer and impaired cell adhesion are shown by the asterisk. Number of cloned placentae examined: 2i/L ES cells #1; n = 8, 2i/L ES cells #2; n = 11, S/L ES cells #8; n = 7, and S/L ES cells #9; n = 9. g, 4n complementation results of 2i/L ES cells and S/L ES cells showing numbers of embryos transferred, full-term pups, and breathing. Images show a GFP-positive all-ES-cell pup (arrow) and an all-ES-cell mouse at 3 weeks of age (right). h, Confirmation of the origin of all-ES-cell pups (pup1 and pup2) by simple sequence length polymorphism (SSLP) using genomic DNA isolated from the original S/L ES cells (#8, XX), pup1, and pup2 in comparison with MSM/Ms and 129X1/SvJ genomic DNA. Genomic DNA from BDF1 mouse was used as control for host blastocyst-derived cells.
Extended Data Figure 7 Loss of gamete-derived methylation is associated with impaired developmental potential of 2i/L ES cells.
a, Doxycycline (Dox)-inducible piggyBac vector containing tetO-Cdx2-IRES-mCherry-EF1-rtTA-IRES-Neo (PB-Cdx2). b, Mean ± s.d. of trophoblast stem cell (TSC)-like colony number after ectopic Cdx2 expression in ES cells. Impaired colony formation was observed in 2i/L ES cells. The number of TSC-like colonies (>300 μm) per 6-well plate is shown (independent experiments, n = 3). c, qRT–PCR analysis for Scml2 in 2i/L ES cells and S/L ES cells. 2i/L ES cells exhibit an increased expression of Scml2. Data are presented as the mean ± s.d. The mean expression level of S/L ES cells (#8) was set to 1 (technical replicates, n = 3). d, qRT–PCR analysis for Scml2 in 2i/L ES cells, 2i/L-S/L ES cells and S/L ES cells. Scml2 expression was repressed in 2i/L-S/L ES cells compared to 2i/L ES cells. Data are presented as the mean ± s.d. The mean expression level of S/L ES cells (#36) was set to 1 (technical replicates, n = 3). e, Maternal Scml2 methylation in 2i/L ES cells (#15, XX), 2i/L-S/L ES cells (#15, XX) and S/L ES cells (#8, XX). CpG sites in which maternal allele has been distinguished were used to determine DNA methylation levels at Scml2 (TSS ± 1,000 bp). Methyl-seq data were used for the analysis. f, Impaired capacity of TSC-like colony formation was partially rescued by serum exposure. Data are mean ± s.d. (independent experiments, n = 3). g, Strategy of knockout for Scml2 using CRISPR–Cas9. gRNA targeting sequence is underlined, and the PAM sequence is labelled in green. 2i/L ES cells (#2, XX) were transfected with plasmids expressing CRISPR–Cas9 and gRNA in 2i/L culture condition. h, Genetic disruption of Scml2 partially rescued the impaired colony formation in 2i/L ES cells. i, DNA methylation analysis for 2i/L-MEFs. 4n 2i/L-MEFs and 2n 2i/L-MEFs (E14.5) exhibit similar methylation levels. ICR methylation data of 2n 2i/L-MEFs is the same as Extended Data Fig. 5b. j, Microarray analysis shows aberrant global gene expression in 4n 2i/L-MEFs. Note that the most affected genes often include imprinted genes. Green lines are fold change lines with a value of 2.0. Representative imprinted genes with altered expression are indicated in the scatter plot.
Extended Data Figure 8 Derivation of ground-state ES cells maintaining ICR methylation.
a, A qRT–PCR analysis for representative Mek target genes (Dusp6 and Cldn7) in ES cells under various PD concentrations and 3μM CHIR. Attenuated repression of these genes was observed until the concentration decreased to 0.2 μM. Data are presented as the mean ± s.d. The mean expression level of S/L ES cells was set to 1 (technical replicates, n = 3). b, Representative morphology of ES cells under various culture conditions. Scale bars, 100 μm. c, Effect of ES-cell-derivation conditions on phosphorylation of ERK1/2 by western blot. d, The relative expressions of Mek target genes in ES cells27. The mean expression level of S/L ES cell lines was set to 1. Two independent clones at p3 were analysed for each culture condition. e, The relative expressions of Gsk target genes in ES cells27 established under various culture conditions. The repression of Gsk target genes is similarly observed in 2i/L, t2i/L and a2i/L ES cells. Two independent clones at p3 were analysed for each culture condition. The mean expression level at S/L ES cell lines was set to 1. f, Hierarchical clustering analysis by RNA-seq data depicts t2i/L ES cells and a2i/L ES cells as having similar expression signatures with 2i/L ES cells (independent ES cell clones; n = 2 for each condition and sex). All expressed genes including X-linked genes were used for this analysis. g, DNA methylation percentage of the whole genome and ICR measured by WGBS in male ES cell lines established under various culture conditions. h, Conventional bisulfite sequencing at Nespas-Gnasxl and Snrpn ICR in multiple clones. Parental alleles were distinguished by SNPs (129 and C57BL6). Note that t2i/L ES cells and a2i/L ES cells tend to retain ICR methylation at maternal allele. i, Allelic expression analysis of Snrpn gene in female 2i/L ES cells, t2i/L ES cells and a2i/L ES cells. RNA-seq analysis reveals that Snrpn tends to be expressed monoallelically in t2i/L ES cells and a2i/L ES cells, whereas the biallelic expression is observed in 2i/L ES cells. j, Efficiency of 4n complementation using 2i/L, t2i/L and a2i/L ES cells. k, Images show all-ES-cell pups originated from female a2i/L ES cells. The origin of all-ES-cell pups was confirmed by SSLP. Genomic DNA from the BDF1 mouse was used as a control for host blastocyst-derived cells.
Extended Data Figure 9 Decreased levels of Uhrf1 protein in female ES cells.
a, Reduction of ICR methylation in female S/L ES cells after prolonged culture. Methylation data of S/L ES cells #8 at p3 are the same as Fig. 1d. b, ICR methylation level in female t2i/L ES cells at p3 and p15. Methylation data of t2i/L ES cells #t1 and #t3 at p3 are the same as Fig. 4d. c, Western blotting of DNA methyltransferases. Dnmt3a and Dnmt3b levels are lower in female ES cells. Dnmt3b was strongly repressed in 2i/L, t2i/L and a2i/L culture conditions. The Dnmt1 level is comparable between male and female ES cells. d, Western blotting for Uhrf1 in multiple S/L ES cells and MEFs and for Uhrf1, Dnmt3a and Dnmt3b in XX/XO/XY S/L ES cells. Note that Uhrf1 protein levels are decreased in female ES cells but not in female MEFs and XO ES cells, suggesting that two active X chromosomes are associated with the decreased Uhrf1 protein level. e, Karyotype analysis by Q-band in S/L ES cells (#s100, p3) unexpectedly revealed 39 chromosomes containing a single X chromosome but lacking a Y chromosome. Of 20 cells, 15 exhibited loss of the Y chromosome. f, Transcript levels of Uhrf1 in ES cells by RNA-seq. Uhrf1 is similarly expressed in both male and female ES cells regardless of culture conditions. g, Ectopic expression of Uhrf1 by Dox-inducible piggyBac vector in female S/L ES cells. mCherry signal confirmed successful induction of the transgene. Scale bars, 100 μm. Uhrf1 protein is not increased in female ES cells by Dox treatment (2 μg ml−1).
Extended Data Figure 10 Increased global DNA methylation and loss of X chromosome in female 2i/L and a2i/L ES cells after prolonged culture.
a, Effect of prolonged culture on global DNA methylation in 2i/L and a2i/L ES cells. Global DNA methylation is increased in female 2i/L and a2i/L ES cells after prolonged culture (p15). Methylation data of female a2i/L ES cells (#a4, #a18) at p3, female 2i/L ES cells (#c3, #c4) at p3, male a2i/L ES cells (#a1, #a3) at p3 and male 2i/L ES cells (#c2, #c6) at p3 were the same as Fig. 4d and Extended Data Fig. 8g. b, pERK of female 2i/L and a2i/L ES cells at p15 are similarly repressed compared with their identical clones at p3. c, Prolonged passage results in an increment of Dnmt3b and Uhrf1 protein levels in female 2i/L and a2i/L ES cells but not in male ES cells. d, CGH analysis depicting partial loss of the X chromosome in female 2i/L and a2i/L ES cells after prolonged culture (p15). Gain of chromosome 8 is observed in both male and female 2i/L ES cells, but not in a2i/L ES cells. e, Mapping efficiency of WGBS reads on the X chromosome in early passage (p3) and late passage (p15) female 2i/L and a2i/L ES cells (#c3, XX; #c4, XX; #a4, XX; #a18, XX). Consistent with the loss of the X chromosome in the CGH analysis, decreased mapping of WGBS reads on the X chromosome is observed in female 2i/L and a2i/L ES cells at p15. Note that the identical ES cell lines at p3 do not display decreased mapping efficiency on the X chromosome, indicating that the female 2i/L and a2i/L ES cell lines have lost the X chromosome during prolonged culture. XX, XO, XY S/L ES cells are shown as controls of X chromosome mapping.
Supplementary information
Supplementary Data
This file contains uncropped gel image data from figures 4g and extended data figures 4c, 6h, 8c, 8k, 9c, 9d, 9g, 10b and 10c. (PDF 839 kb)
Rights and permissions
About this article
Cite this article
Yagi, M., Kishigami, S., Tanaka, A. et al. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature 548, 224–227 (2017). https://doi.org/10.1038/nature23286
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23286
This article is cited by
-
Amino acid intake strategies define pluripotent cell states
Nature Metabolism (2024)
-
Derivation of embryonic stem cells from wild-derived mouse strains by nuclear transfer using peripheral blood cells
Scientific Reports (2023)
-
Optimization of Cas9 activity through the addition of cytosine extensions to single-guide RNAs
Nature Biomedical Engineering (2023)
-
Enhanced cultivation of chicken primordial germ cells
Scientific Reports (2023)
-
Sex-biased and parental allele-specific gene regulation by KDM6A
Biology of Sex Differences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.