Abstract
Although long non-coding RNAs (lncRNAs) are non-protein-coding transcripts by definition, recent studies have shown that a fraction of putative small open reading frames within lncRNAs are translated1,2,3. However, the biological significance of these hidden polypeptides is still unclear. Here we identify and functionally characterize a novel polypeptide encoded by the lncRNA LINC00961. This polypeptide is conserved between human and mouse, is localized to the late endosome/lysosome and interacts with the lysosomal v-ATPase to negatively regulate mTORC1 activation. This regulation of mTORC1 is specific to activation of mTORC1 by amino acid stimulation, rather than by growth factors. Hence, we termed this polypeptide ‘small regulatory polypeptide of amino acid response’ (SPAR). We show that the SPAR-encoding lncRNA is highly expressed in a subset of tissues and use CRISPR/Cas9 engineering to develop a SPAR-polypeptide-specific knockout mouse while maintaining expression of the host lncRNA. We find that the SPAR-encoding lncRNA is downregulated in skeletal muscle upon acute injury, and using this in vivo model we establish that SPAR downregulation enables efficient activation of mTORC1 and promotes muscle regeneration. Our data provide a mechanism by which mTORC1 activation may be finely regulated in a tissue-specific manner in response to injury, and a paradigm by which lncRNAs encoding small polypeptides can modulate general biological pathways and processes to facilitate tissue-specific requirements, consistent with their restricted and highly regulated expression profile.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011)
Kim, M. S. et al. A draft map of the human proteome. Nature 509, 575–581 (2014)
Slavoff, S. A. et al. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64 (2013)
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007)
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005)
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012)
Pauli, A. et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343, 1248636 (2014)
Kondo, T. et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329, 336–339 (2010)
Magny, E. G. et al. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341, 1116–1120 (2013)
Anderson, D. M. et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606 (2015)
Nelson, B. R. et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275 (2016)
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011)
Humphries, W. H., IV, Szymanski, C. J. & Payne, C. K. Endo-lysosomal vesicles positive for Rab7 and LAMP1 are terminal vesicles for the transport of dextran. PLoS One 6, e26626 (2011)
Rebsamen, M. et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481 (2015)
Zoncu, R. et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 (2011)
Bar-Peled, L. & Sabatini, D. M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24, 400–406 (2014)
Sancak, Y. et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010)
Sancak, Y. et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008)
Bar-Peled, L., Schweitzer, L. D., Zoncu, R. & Sabatini, D. M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012)
Bentzinger, C. F. et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424 (2008)
Ge, Y. et al. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am. J. Physiol. Cell Physiol. 297, C1434–C1444 (2009)
Pereira, M. G. et al. Leucine supplementation accelerates connective tissue repair of injured tibialis anterior muscle. Nutrients 6, 3981–4001 (2014)
Zhang, P. et al. mTOR is necessary for proper satellite cell activity and skeletal muscle regeneration. Biochem. Biophys. Res. Commun. 463, 102–108 (2015)
Li Chew, C. et al. In vivo role of INPP4B in tumor and metastasis suppression through regulation of PI3K-AKT signaling at endosomes. Cancer Discov. 5, 740–751 (2015)
Acknowledgements
We thank members of the P.P.P. laboratory, C. C. Dibble for critical discussions and Cell Signaling Technology for generation of SPAR antibodies. A.M. was supported by a postdoctoral fellowship from JSPS, The Uehara Memorial Foundation and The Naito Foundation. This work was supported in part by NIH grant R01 CA082328 and R35 CA197529 to P.P.P. and JST and PREST to A.M.
Author information
Authors and Affiliations
Contributions
A.M. conceived the project, designed and performed most experiments, interpreted the results, and co-wrote the manuscript. A.P. performed muscle experiments. M.M. performed mass spectrometry. R.Y. performed informatic analysis. J.F. performed histology and immunoblotting experiments. E.M. analysed whole-genome sequencing data. A.S. and K.I.N. supervised experimental designs. J.G.C. supervised experimental designs and co-wrote the manuscript. P.P.P. conceived the project, supervised experimental designs, interpreted results, and co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks K.-L. Guan, M. Rüegg and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
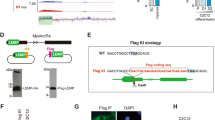
Extended Data Figure 1 Characterization of the LINC00961 RNA and encoded polypeptide.
a, qPCR analysis of HPRT, circHIPK3, and LINC00961 using mRNA from HeLa cells reverse transcribed with either random or oligo-dT primers. Data are mean ± s.d. (n = 3). b, Representative ribosome profile. c, HeLa cell lysates were fractionated to collect free 40/60S subunit, monosome, di/trisome and polysome fractions by sucrose gradient centrifugation as shown in b. HPRT, circHIPK3, and LINC00961 were extracted from these fractions and quantified by qPCR. d, MS/MS spectra from SKOV3 cell lysate of an endogenous polypeptide encoded by LINC00961 (left), and synthetic peptide MS/MS spectra confirming spectral pattern of the endogenous peptide (right). e, f, qPCR analysis of LINC00961 using RNA isolated from human cell lines (e) and human tissues (f). Normalized data are expressed relative to the value for HeLa cells and bone marrow, respectively. Data shown as mean ± s.d. of technical triplicates. g, qPCR analysis of the mouse homologue 5430416O09Rik in mouse primary tissues. Normalized data are expressed relative to the value for cerebrum. Data shown as mean ± s.d. of technical triplicates. h, Nucleotide and amino acid sequences of human LINC00961-encoded polypeptide. AUG codons and transmembrane region are highlighted in red. i, Schematic representation of all LINC00961 constructs used in this study. All expression constructs included endogenous 5′ and 3′ UTR regions, but only CDSs are shown here for simplicity. j, qPCR analysis of the LINC00961 RNA from HEK293T cells expressing LINC00961 Flag KI1, Flag KI1-ΔATG1, Flag KI1-ΔATG2 and Flag KI1-ΔATG1+2. Normalized data are expressed relative to the value for Flag KI1. Data shown as mean ± s.d. (n = 3). k, l, Cycloheximide (CHX) chase for Flag KI1-ΔATG1 and Flag KI1-ΔATG2 (d), quantified in e. Data shown as mean ± s.d. (n = 3). For gel source images, see Supplementary Fig. 1.
Extended Data Figure 2 Topology and localization of the LINC00961-encoded polypeptide.
a, Total, soluble (Sup.) and membrane (Pellet) fractions from HEK293T cells stably expressing Flag KI1 were subjected to immunoblot analysis to evaluate membrane association. b, Homogenates of HEK293T cells stably expressing Flag KI1 were incubated with or without 0.1 M Na2CO3, 0.1 M NaOH or 1 M NaCl and subsequently centrifuged to yield the soluble fraction (Sup.) and membrane fraction (Pellet). Fractions were then subjected to immunoblot analysis to evaluate membrane integration. c, Schematic representation of the membrane topology of calnexin, GM130, and the Flag KI1 LINC00961-encoded polypeptide. The N-terminal epitope of calnexin recognized by the anti-calnexin antibody is indicated. d, Membrane fractions from HEK293T cells stably expressing Flag KI1 were incubated in the absence or presence of proteinase K and/or Triton X-100 and were subjected to immunoblot analysis. e, Immunofluorescence images for Flag KI1, EEA1 (localized to early endosome) and catalase (localized to peroxisome) in HeLa cells. A representative image from at least three fields of view recorded is shown. Scale bar, 5 μm. f, Immunofluorescence images (from two fields of view imaged) demonstrating co-localization of Flag KI1 and LAMP1 in PC3 cells. Scale bar, 2.5 μm. g, Schematic representation of chimaeric CD8 and LINC00961-encoded polypeptide (ORF1) fusion constructs used to determine the region responsible for lysosomal localization. h–j, HeLa cells were transfected with full-length CD8 (CD8(WT)) (h), with a chimaeric CD8 construct with its transmembrane domain replaced with the LINC00961 polypeptide transmembrane domain (CD8(N)-ORF1(TM)-CD8(C)) (i), or with a chimaeric construct in which the cytoplasmic region of CD8 was replaced with the LINC00961 polypeptide cytoplasmic region (CD8(N and TM)-ORF1(C)) (j). Cells were subsequently subjected to immunofluorescence staining to detect CD8 (Alexa-488) and LAMP1 (Alexa-546). A representative image from at least two fields of view recorded is shown. Scale bar, 5 μm. k, qPCR analysis for knockdown of the LINC00961 RNA in HeLa cells treated with siControl, siLINC00961_1 and siLINC00961_2. Normalized data are expressed relative to the value for siControl. Data shown as mean ± s.d. (n = 3). l, HeLa cell lysates were subjected to immunoprecipitation with IgG control or with antibody no. 2 against LINC00961-encoded polypeptide followed by immunoblotting for ATP6V0A1, ATP6V0A2 and ORF1 using antibody no. 2. For gel source images, see Supplementary Fig. 1.
Extended Data Figure 3 v-ATPase localization, assembly and lysosomal functions are unaffected by the LINC00961-encoded polypeptide.
a, Localization of v-ATPase subunits was evaluated across membrane fractions by sucrose density gradient fractionation of lysates from HEK293T cells transduced with Mock, wild-type LINC00961 or mutant LINC00961 (ΔATG1+2). v-ATPase localization was characterized by immunoblot analysis for the ATP6V0A1, ATP6V0A2, ATP6V0D1, and ATP6V1A subunits, while immunoblot analysis for LAMP2, calnexin and EEA1 served as controls. b, c, Soluble (Sup.) and membrane (Pellet) fractions from HEK293T cells stably expressing Mock, wild-type LINC00961 or mutant LINC00961 (ΔATG1+2) were immunoblotted for the V1 domain subunit ATP6V1A, the V0 domain subunit ATP6V0D1, and LAMP2 (b). The relative steady-state ratio of ATP6V1A/ATP6V0D1 in pellet fractions was used as a measure of v-ATPase assembly (c). Treatment with chloroquine promotes v-ATPase assembly and serves as a positive control to measure changes in assembly. Data shown as mean ± s.d. (n = 3). d, HEK293T cells were treated for 3 h with DMSO or 200 nM of the v-ATPase inhibitor Bafilomycin A1 (BafA1), an inhibitor of proton pump activity that increases lysosomal pH. Lysosomal pH was monitored by flow cytometry using Lysosensor DND-189 (left), and the mean fluorescence intensity (MFI) plotted to illustrate decreased acidification (right). Data shown as mean ± s.d. (n = 3). e, HEK293T cells stably expressing empty vector (Mock) or wild-type LINC00961 were used to examine the impact of the LINC00961 polypeptide on lysosomal pH. Cells were analysed by flow cytometry as in d, and MFI was plotted. Data are mean ± s.d. (n = 3). f, g, The activity of cathepsin L (f) and cathepsin K (g) was measured using a Magic Red Cathepsin Assay Kit and quantified. HEK293T cells treated with DMSO or 200 nM BafA1 (positive control) for 3 h (left), or HEK293T cells stably expressing control and wild-type LINC00961 (right), were analysed. Data shown as mean ± s.d. (n = 3). h–k, Lysosomal morphology was analysed by immunostaining for LAMP2. HEK293T cells were treated with DMSO or 20 nM BafA1 for 16 h. A representative image from at least four fields of view recorded is shown (h) and lysosomal diameters were measured and quantified (i). Similarly, HEK293T cells stably expressing vector control (Mock) or wild-type LINC00961 constructs were analysed with or without amino acid deprivation for 1 h. A representative image from at least six fields of view recorded is shown (j) and their lysosomal diameters were measured and quantified (k). n = 100 loci per condition. Scale bars, 5 μm. ***P < 0.001, ****P < 0.0001; Student’s t-test (d–g, i), or one-way (c) or two-way (k) ANOVA followed by Tukey’s test. For gel source images, see Supplementary Fig. 1.
Extended Data Figure 4 mTORC1 regulation by the LINC00961-encoded polypeptide.
a, Current model of mTORC1 activation and signalling. This model highlights our current understanding of how mTORC1 is recruited to, and activated at, the lysosome. Rag proteins promote recruitment of mTORC1 to the lysosome where it can be activated by Rheb. Amino acid stimulation releases the Ragulator from the v-ATPase, whereby it interacts with Rags to facilitate mTORC1 recruitment and subsequent activation. Rag proteins can also be regulated through additional mechanisms involving the amino acids leucine and arginine as illustrated. The LINC00961-encoded polypeptide (SPAR) acts at the level of the v-ATPase to promote and stabilize the interaction between the v-ATPase, Ragulator and Rags to inhibit mTORC1 recruitment and activation at the lysosome, even in the presence of amino acid stimulation. b, Cell proliferation analysis as analysed by MTT assay in HEK293T cells infected with mock, wild-type LINC00961 and the ΔATG1+2 mutant. Data are mean ± s.d. from technical triplicates. c, S6K1 and S6 phosphorylation status remain unchanged in HEK293T cells, stably expressing wild-type LINC00961 and the ΔATG1+2 mutant under steady state conditions. d, S6K1 phosphorylation in PC3 (upper panel) and HeLa cells (lower panel) stably expressing wild-type LINC00961 and ΔATG1+2 mutants, deprived of amino acids for 1 h and stimulated with amino acids for indicated times. e, f, mTORC1 activation as evaluated by S6K1, S6 and 4EBP phosphorylation in HEK293T cells (e), and the relative ratio of phospho-S6K to total S6K (f), for stably expressing wild-type LINC00961, ΔATG2, ΔATG1 and ΔATG1+2 mutants, deprived of amino acids for 1 h and stimulated with amino acids for 10 min. Data shown as mean ± s.d. (n = 3) g, mTORC1 activation as evaluated by S6K1 and S6 phosphorylation in HEK293T cells stably expressing wild-type LINC00961 and the Flag KI1 mutant. Cells were deprived of amino acids for 1 h and subsequently stimulated with amino acids for 10 min. h, Schematic representation of the LINC00961 ORF1 and Mito-ORF1-Flag, which contains two consecutive mitochondrial targeting signals derived from the cytochrome c oxidase subunit VIII fused to the N terminus of LINC00961 ORF1 and a Flag tag at its C terminus (left), and a representative immunofluorescence image, from five fields of view recorded, demonstrating co-localization in HeLa cells of Mito-ORF1-Flag with Mitotracker Red, a marker for mitochondria (right). Scale bar, 10 μm. i, j, mTORC1 activation as evaluated by S6K1 and S6 phosphorylation in HEK293T cells (i), quantified in j, for cells stably expressing Mito-ORF1 (without Flag tag) and wild-type LINC00961, deprived of amino acids for 1 h and stimulated for 10 min. Data shown as mean ± s.d. (n = 3). ***P < 0.001. One-way ANOVA followed by Tukey’s test (j). For gel source images, see Supplementary Fig. 1.
Extended Data Figure 5 The LINC00961-encoded polypeptide fails to influence signalling independent of amino acid stimulation.
a, b, Phosphorylation status of S6K1, S6 and 4EBP in HEK293T cells stably expressing vector control (Mock), wild-type LINC00961 or mutant LINC00961 (ΔATG1+2), deprived of serum for 24 h and stimulated with 1 μM insulin for the times indicated (a), and quantitative analysis of relative level of phospho-S6K to total S6K 30 min after insulin stimulation (b). Data shown as mean ± s.d. (n = 3). c–e, S6K1 and S6 and phosphorylation status in HEK293T cells stably expressing vector control (Mock) or wild-type LINC00961 after complete amino acid depletion, leucine depletion or arginine depletion for 1 h and stimulation for 10 min with the respective amino acids as shown (c), and quantitative analysis of relative level of phospho-S6K to total S6K after leucine (d) or arginine (e) stimulation. Data shown as mean ± s.d. (n = 3). f–h, Phosphorylation status of AKT (S473) and ERK1/2 in HEK293T cells stably expressing vector control (Mock) or wild-type LINC00961, deprived of serum for 24 h and stimulated with 1 μM insulin or 10 ng ml−1 EGF for 10 min (f), and quantitative analysis of relative level of phospho-AKT to total AKT after insulin stimulation (g), or phospho-ERK to total ERK after EGF stimulation (h). Data shown as mean ± s.d. (n = 3). Student’s t-test (d, e, g, h) or one-way ANOVA followed by Tukey’s test (b). For gel source images, see Supplementary Fig. 1.
Extended Data Figure 6 Characterization of mouse Spar.
a, Nucleotide and amino acid sequence of the mouse homologue 5430416O09Rik-encoded polypeptide. AUG codon and transmembrane region are highlighted in red. b, Polypeptide identity between human and mouse homologues. Identical amino acids are indicated in red. c, Immunoblotting with anti-Flag of HEK293T cell lysates transfected with expression constructs for full-length 5430416O09Rik (mWT), C-terminal Flag knockin (mFlag KI), or C-terminal Flag KI with deletion of the initiation ATG (mFlag KI-ΔATG). d, e, mTORC1 activation as evaluated by S6K1 and S6 phosphorylation in HEK293T cells (d) and mouse C2C12 cells (e) stably expressing mouse 5430416O09Rik (mWT) or the mΔATG mutant. Cells were deprived of amino acids for 1 h and subsequently stimulated with amino acids for the indicated times. f, Membrane (Pellet) and soluble (Sup.) fractions from mouse skeletal muscle were subjected to immunoprecipitation of Spar, followed by immunoblotting for Spar and Lamp1. g, h, qPCR analysis of mouse Atp6v0a1 (g) and Atp6v0a2 (h) in mouse primary tissues. Normalized data are expressed relative to the value for cerebrum. Data shown as mean ± s.d. of technical triplicates. i, Immunoprecipitation of murine Spar from skeletal muscle lysates followed by immunoblotting for Atp6v0a1 and Atp6v0a2. For gel source images, see Supplementary Fig. 1.
Extended Data Figure 7 Generation and characterization of Spar-deficient mice.
a, sgRNA sequences and ssDNA oligo sequence used for CRISPR/Cas9-mediated homologous recombination to generate Spar-deficient mice. ATG initiation codon and SacII restriction enzyme site are highlighted in red. A SacII restriction enzyme site close to the ΔATG was mutated in mutant mice to distinguish the mutant allele from the wild-type allele after PCR. b, Analysis of genomic tail DNA by PCR and SacII treatment for mice of the indicated genotypes at 4 weeks of age. c, Sanger sequencing result of Spar locus from genomic DNA of the Spar–/– mouse. d, qRT–PCR analysis of the 5430416O09Rik RNA in tibialis anterior (TA) muscles from Spar+/+ and Spar–/– mice. Data shown as mean ± s.d. (n = 4). e. Confirmation of the Spar mutation by WGS. IGV images show the ΔATG homozygous mutation within the second exon of Spar. f, g, Sanger sequencing result for SNPs of Sfi1-exon5 (f) and Sfi1-exon13 (g) loci from genomic DNA of the Spar–/– mouse. h, Frequency of genotypes produced from Spar+/– mouse intercrosses. Numbers in parentheses indicate the expected number by Mendelian ratio. i, Bodyweight of Spar+/+ and Spar–/– mice at 8 weeks of age. Data shown as mean ± s.d. (n = 9). j, Localization of v-ATPase subunits in tibialis anterior muscle from Spar+/+ and Spar–/– mice was evaluated across membrane fractions by sucrose density gradient fractionation. v-ATPase localization was characterized by immunoblot analysis for the Atp6v0a1, Atp6v0a2, Atp6v0d1, and Atp6v1a subunits, while immunoblot analysis for Lamp1 served as control. k, l, Soluble (Sup.) and membrane (Pellet) fractions from tibialis anterior muscle of Spar+/+ and Spar–/– mice were immunoblotted for the V1 domain subunit Atp6v1a, the V0 domain subunit Atp6v0d1, and Lamp1 (k). The relative steady-state ratio of Atp6v1a to Atp6v0d1 in pellet fractions was used as a measure of v-ATPase assembly (l). Data shown as mean ± s.d. (n = 3). m, Myoblasts derived from Spar+/+ mice were treated with DMSO or 200 nM of the v-ATPase inhibitor Bafilomycin A1 (BafA1) for 3 h. Lysosomal pH was monitored by flow cytometry using Lysosensor DND-189 and mean fluorescence intensity (MFI) was plotted. Data shown as mean ± s.d. (n = 3). n, Myoblasts derived from Spar+/+ and Spar–/– mice were used to examine the impact of Spar on lysosomal pH. Cells were analysed by flow cytometry as in m, and MFI was plotted. Data shown as mean ± s.d. (n = 3). o, p, Lysosomal morphology was analysed by immunostaining for Lamp1 in tibialis anterior muscle from Spar+/+ and Spar–/– mice at 8 weeks of age. Representative image (from ten immunofluorescence images recorded per condition) (o) and lysosomal diameters were measured and quantified (p). n = 150 loci per condition. Scale bar, 10 μm. ***P < 0.001; Student’s t-test (d, l–n, p) or two-way ANOVA followed by Tukey’s test (i). For gel source images, see Supplementary Fig. 1.
Extended Data Figure 8 Regulation of muscle regeneration by mTORC1 and Spar.
a, A schematic outlining the experimental timeline to study the role of mTORC1 in muscle regeneration. Rapamycin (2 mg per kg per day) or vehicle control was administered daily by intraperitoneal (i.p.) injection, with treatment commencing the day before CTX administration. b, c, S6K1 and S6 phosphorylation status in regenerating tibialis anterior muscle from mice treated with rapamycin or vehicle control 4 days after CTX administration (b), and quantitative analysis of relative levels of phospho-S6K to total S6K (c). Data shown as mean ± s.d. (n = 3). d, Per cent change in weight of regenerating tibialis anterior muscle compared with that of uninjured lateral control muscle. Mice were treated with rapamycin or vehicle control one day before CTX administration and muscles analysed 14 days after CTX administration. Data shown as mean ± s.d. (n = 4). e, f, H&E staining of regenerating tibialis anterior muscle from mice treated with vehicle control or rapamycin. Representative images (from at least five fields of view per mouse) (e), and quantification of mean area of regenerating cells with central nuclei (f). Data shown as mean ± s.d. (n = 4). Scale bar, 50 μm. g, Bodyweight of mice treated with vehicle control or rapamycin 14 days after CTX administration. Data shown as mean ± s.d. (n = 4). h, 14-day time course of Pax7 and myogenin expression by qRT–PCR analysis in tibialis anterior muscle from Spar+/+ mice post-CTX administration. Data shown as mean ± s.d. (n = 3). i, A schematic outlining the experimental timeline to study the role of Spar in muscle regeneration. j, k, Mice were placed on control or leucine-free diet one day before CTX administration, with body weight (j) and food intake (k) of Spar+/+ and Spar–/– mice analysed 7 days after CTX administration. Similar conditions were used in l–o. l, Uninjured lateral control tibialis anterior muscle weight from Spar+/+ and Spar–/– mice. m, Uninjured lateral control tibialis anterior muscle weights normalized by total body weight in Spar+/+ and Spar–/– mice. n, Regenerating tibialis anterior muscle weight from Spar+/+ and Spar–/– mice. o, Regenerating tibialis anterior muscle weights normalized by total body weight from Spar+/+ and Spar–/– mice. Data shown as mean ± s.d. (n = 5 for control diet, n = 3 for leucine-free diet, for j, l–o; n = 4 for k). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Student’s t-test (c, d, f, g) or two-way ANOVA followed by Tukey’s test (j–o). For gel source images, see Supplementary Fig. 1.
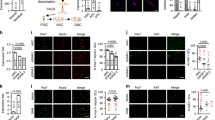
Extended Data Figure 9 Characterization of enhanced regeneration in Spar-deficient tibialis anterior muscle.
a, b, The number of regenerating myofibres with central nuclei (a), and total area of regenerating myofibres with central nuclei (b) in regenerating tibialis anterior muscles 7 days after CTX administration in Spar+/+ and Spar–/– mice on control diet or leucine-free diet. Data shown as mean ± s.d. (n = 5 for control diet, n = 3 for leucine-free diet). c, d, Percentage of myofibres with central nuclei from a and b analysed according to cell size. Comparisons for Spar+/+ and Spar–/– mice on control diet (c) and leucine-free diet (d) are shown. Data shown as mean ± s.d. (n = 5 for control diet, n = 3 for leucine-free diet). e, f, H&E staining of uninjured lateral control tibialis anterior muscle from Spar+/+ and Spar–/– mice. Representative images (from at least 10 fields of view per mouse) (e), and mean area of myofibres (f). Data shown as mean ± s.d. (n = 3). Scale bar, 50 μm. g–i, Immunoblot analysis of Pax7 and Myog in injured tibialis anterior muscle lysates 4 days after CTX administration, from Spar+/+ and Spar–/– mice on control or leucine-free diet (g), and quantitative analysis of relative level of Pax7 (h) and Myogenin (i) normalized to Gapdh level. Data shown as mean ± s.d. (n = 3). j–l, Immunofluorescence analysis for Pax7 and laminin (j), Myog and laminin (k) and Pax7 and Ki67 (l) in regenerating tibialis anterior muscle from Spar+/+ and Spar–/– mice 4 days after CTX administration. Representative images (from at least six fields of view per mouse) are shown, and the quantification is shown in Fig. 4i, j (n = 3). Scale bar, 20 μm. m, H&E staining of regenerating tibialis anterior muscle from Spar+/+ and Spar–/– mice 14 days after CTX administration. Representative images (from at least 10 fields of view per mouse) are shown, and percentage of mature cells with peripheral nuclei was quantified in Fig. 4k (n = 3). Scale bar, 50 μm. n, A schematic outlining the experimental timeline to study regeneration of gastrocnemius muscle. o, p, Phosphorylation status of S6K1 and S6 in regenerating gastrocnemius muscle from Spar+/+ and Spar–/– mice 4 days after injury (o), and quantitative analysis of relative level of phospho-S6K/total-S6K (p). Data shown as mean ± s.d. (n = 3). q, Per cent change in weight of regenerating gastrocnemius muscle compared with that of uninjured control lateral muscle. Muscles were removed 7 days after CTX administration. Data shown as mean ± s.d. (n = 4). r, s, H&E staining of regenerating gastrocnemius muscle from Spar+/+ and Spar–/– mice 7 days after CTX administration. Representative images (from at least five fields of view per mouse) (r), and mean area of cells with central nuclei (s). Data shown as mean ± s.d. (n = 4). Scale bar, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Student’s t-test (p, q, s) or two-way ANOVA followed by Tukey’s test (a–d, f, h, i). For gel source images, see Supplementary Fig. 1.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-6, Supplementary References and Supplementary Figure 1, which contains immunoblot source data. (PDF 840 kb)
Supplementary Table 1
This file contains the full list of protein interaction partners for the SPAR polypeptide identified by mass spectrometric analyses. (XLSX 260 kb)
Supplementary Table 2
Whole genome sequencing data summary, this file contains the variant sites (Read depth > 50) observed by whole genome sequencing of three independent Spar KO mice. (XLSX 59 kb)
Rights and permissions
About this article
Cite this article
Matsumoto, A., Pasut, A., Matsumoto, M. et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232 (2017). https://doi.org/10.1038/nature21034
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21034
This article is cited by
-
Micropeptides: potential treatment strategies for cancer
Cancer Cell International (2024)
-
A lncRNA Dleu2-encoded peptide relieves autoimmunity by facilitating Smad3-mediated Treg induction
EMBO Reports (2024)
-
Small open reading frames: a comparative genetics approach to validation
BMC Genomics (2023)
-
A review on the role of long non-coding RNA and microRNA network in clear cell renal cell carcinoma and its tumor microenvironment
Cancer Cell International (2023)
-
Amino acid metabolism regulated by lncRNAs: the propellant behind cancer metabolic reprogramming
Cell Communication and Signaling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.