Abstract
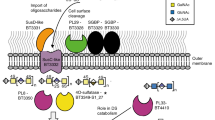
The human large intestine is populated by a high density of microorganisms, collectively termed the colonic microbiota1, which has an important role in human health and nutrition2. The survival of microbiota members from the dominant Gram-negative phylum Bacteroidetes depends on their ability to degrade dietary glycans that cannot be metabolized by the host3. The genes encoding proteins involved in the degradation of specific glycans are organized into co-regulated polysaccharide utilization loci4,5,6,7,8, with the archetypal locus sus (for starch utilisation system) encoding seven proteins, SusA–SusG8,9,10. Glycan degradation mainly occurs intracellularly and depends on the import of oligosaccharides by an outer membrane protein complex composed of an extracellular SusD-like lipoprotein and an integral membrane SusC-like TonB-dependent transporter4,5,6,7,11,12,13. The presence of the partner SusD-like lipoprotein is the major feature that distinguishes SusC-like proteins from previously characterized TonB-dependent transporters. Many sequenced gut Bacteroides spp. encode over 100 SusCD pairs, of which the majority have unknown functions and substrate specificities3,8,14,15. The mechanism by which extracellular substrate binding by SusD proteins is coupled to outer membrane passage through their cognate SusC transporter is unknown. Here we present X-ray crystal structures of two functionally distinct SusCD complexes purified from Bacteroides thetaiotaomicron and derive a general model for substrate translocation. The SusC transporters form homodimers, with each β-barrel protomer tightly capped by SusD. Ligands are bound at the SusC–SusD interface in a large solvent-excluded cavity. Molecular dynamics simulations and single-channel electrophysiology reveal a ‘pedal bin’ mechanism, in which SusD moves away from SusC in a hinge-like fashion in the absence of ligand to expose the substrate-binding site to the extracellular milieu. These data provide mechanistic insights into outer membrane nutrient import by members of the microbiota, an area of major importance for understanding human–microbiota symbiosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A. & Gordon, J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005)
Marchesi, J. R. et al. The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339 (2015)
Koropatkin, N. M., Cameron, E. A. & Martens, E. C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10, 323–335 (2012)
Sonnenburg, E. D. et al. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010)
Rogowski, A. et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 6, http://dx.doi.org/10.1038/ncomms8481 (2015)
Larsbrink, J. et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 (2014)
Cuskin, F. et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169 (2015)
Martens, E. C., Koropatkin, N. M., Smith, T. J. & Gordon, J. I. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284, 24673–24677 (2009)
D’Elia, J. N. & Salyers, A. A. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 178, 7173–7179 (1996)
Reeves, A. R., Wang, G. R. & Salyers, A. A. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179, 643–649 (1997)
Koropatkin, N. M., Martens, E. C., Gordon, J. I. & Smith, T. J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 (2008)
Shipman, J. A., Berleman, J. E. & Salyers, A. A. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182, 5365–5372 (2000)
Noinaj, N., Guillier, M., Barnard, T. J. & Buchanan, S. K. TonB-dependent transporters: regulation, structure, and function. Annu. Rev. Microbiol. 64, 43–60 (2010)
Martens, E. C. et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221 (2011)
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008)
Bolam, D. N. & Koropatkin, N. M. Glycan recognition by the Bacteroidetes Sus-like systems. Curr. Opin. Struct. Biol. 22, 563–569 (2012)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005)
Kumar, S. & Engelberg-Kulka, H. Quorum sensing peptides mediating interspecies bacterial cell death as a novel class of antimicrobial agents. Curr. Opin. Microbiol. 21, 22–27 (2014)
Mohammad, M. M., Howard, K. R. & Movileanu, L. Redesign of a plugged β-barrel membrane protein. J. Biol. Chem. 286, 8000–8013 (2011)
Koropatkin, N., Martens, E. C., Gordon, J. I. & Smith, T. J. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry 48, 1532–1542 (2009)
Shultis, D. D., Purdy, M. D., Banchs, C. N. & Wiener, M. C. Outer membrane active transport: structure of the BtuB:TonB complex. Science 312, 1396–1399 (2006)
Pawelek, P. D. et al. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science 312, 1399–1402 (2006)
Jordan, L. D. et al. Energy-dependent motion of TonB in the Gram-negative bacterial inner membrane. Proc. Natl Acad. Sci. USA 110, 11553–11558 (2013)
Celia, H. et al. Structural insight into the role of the Ton complex in energy transduction. Nature 538, 60–65 (2016)
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 (2004)
Sheldrick, G. M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D 66, 479–485 (2010)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997)
Vagin, A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Buchanan, S. K. et al. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6, 56–63 (1999)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000)
The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC
Schwarz, G., Danelon, C. & Winterhalter, M. On translocation through a membrane channel via an internal binding site: kinetics and voltage dependence. Biophys. J. 84, 2990–2998 (2003)
Montal, M. & Mueller, P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl Acad. Sci. USA 69, 3561–3566 (1972)
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013)
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010)
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012)
Pothula, K. R., Solano, C. J. & Kleinekathöfer, U. Simulations of outer membrane channels and their permeability. Biochim. Biophys. Acta 1858 (7 Pt B), 1760–1771 (2016)
Hess, B., Bekker, H., Berendsen, H. J. C. & Johannes, G. E. M. F. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997)
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5, Unit 6 (2006)
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008)
Coyne, M. J. et al. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol. Microbiol. 88, 772–783 (2013)
Holm, L. & Rosenström, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010)
Acknowledgements
We would like to thank J. Gray for B. theta outer membrane protein identification, R. Lewis for critical reading of the manuscript and S. Buchanan for the pET9 expression vector. We thank O. Davies for help with SEC–MALS analysis. We are also indebted to the staff at beamlines I24, I04 and I03 of the Diamond Light Source UK for beam time (proposal mx9948) and assistance with data collection. A.J.G. acknowledges support from the Barbour Foundation. S.B.P. was funded by EU FP7-PEOPLE-2013-ITN Translocation network Nr. 607694. The research of K.R.P., U.K., M.W. and B.v.d.B. has received support from the Innovative Medicines Initiatives Joint Undertaking under Grant Agreement No. 115525, resources which are composed of financial contributions from the European Union’s seventh framework programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations companies in-kind contribution.
Author information
Authors and Affiliations
Contributions
A.J.G. expressed, purified and crystallized proteins and carried out western blot analysis. K.R.P. and U.K. performed molecular dynamics simulations. S.P.B. and M.W. performed and supervised electrophysiology experiments, respectively. D.S.C. and C.V.R. carried out mass-spectrometry analyses. A.B. collected crystallography data and managed the Newcastle Structural Laboratory. H.Z. expressed and purified BT1762 and performed ITC measurements. S.J.F. and A.B. determined the structure of BT1762. D.N.B. designed research and assisted in writing the paper. B.v.d.B. crystallized proteins and solved crystal structures, designed research and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks B. Henrissat, S. White and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 High abundance of SusCD complexes in B. theta outer membrane.
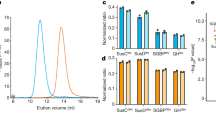
a, SDS–PAGE gel of total outer membranes from E. coli (lanes 1, 2, 5, 6) and B. theta (lanes 3, 4, 7, 8). Rich medium, even lanes; minimal medium, odd lanes. Each lane contains approximately 10 μg protein. Samples 5–8 were boiled. ‘P’ denotes E. coli trimeric porins OmpF/C, which migrate at their monomeric molecular weights (around 35 kDa) only after boiling. Note the relative lack of small-molecule outer membrane diffusion channels (around 30–50 kDa) in B. theta (lanes 7, 8) and the low levels of large outer membrane proteins including TBDTs (70–120 kDa) in E. coli (lanes 5, 6). Purified BT2261–64 complex is shown in lanes 9 (non-boiled) and 10 (boiled). b, Representative ion-exchange chromatogram from three separate experiments of B. theta total outer membrane proteins separated on Resource-Q (6 ml; pH 7.5) after extraction in LDAO (Methods). Peaks A and B were further purified by gel filtration. c, SDS–PAGE gel of purified SusCD complexes from peaks A and B. Numbered gel bands were excised and subjected to identification by mass spectrometry. d, SDS–PAGE gel of purified BT1762–BT1763 complex before (asterisk) and after boiling. The SusCD complexes are highly stable and remain intact in 2% SDS.
Extended Data Figure 2 X-ray crystal structures of the small lipoproteins BT2261 and BT2262.
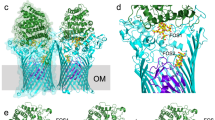
a, Stereo cartoon of BT2261 within the BT2261–64 complex with rainbow colouring (blue; N terminus). BT2261 is O-glycosylated at Ser117, consistent with the presence of the Bacteroidetes glycosylation motif D-(S/T)-(A/L/V/I/M/T)51. Ser117 is shown as a stick model. Fo − Fc density within 20 Å of Ser117 is shown as a green mesh contoured at 3.0σ. Three to four sugar moieties can be observed bound to Ser117. b, Stereo cartoon of soluble BT2262 with rainbow colouring. The protein consists of an N-terminal Ig-like domain and a C-terminal eight-stranded β-barrel. The functions of BT2261 and BT2262 are not clear, but both contain a small C-terminal eight-stranded β-barrel that displays structural similarity to lipid binding domains as judged by DALI52. For BT2262, only one copy with a poorly ordered C-terminal domain is visible in the triclinic structure. Analogous to BT2263, the N-terminal segments of BT2261/BT2262 that lead to the lipid anchors on the N-terminal cysteine residues are visible in the electron density; they are closely associated with SusC and do not appear to be flexible. Structures were determined using data obtained from a single crystal in each case.
Extended Data Figure 3 The oligomeric nature of SusCD complexes is not a consequence of crystal packing.
a–c, Cartoon side views of BT2261–64 complexes rotated by 90° for space groups P1 (a), P212121 (b) and SeMet P21 (c). d, Cartoon side view of BT1762–BT1763 (P212121). The protein backbones are coloured on the same scale by their B-factors (blue; 20 Å2, red; 130 Å2). The grey bars indicate the hydrophobic phase of the outer membrane. Structures were determined using data obtained from a single crystal in each case.
Extended Data Figure 4 BT2261–64 and BT1762–BT1763 form oligomeric complexes.
a, Mass spectrum of BT2261–64 shows two prominent masses corresponding to an octamer and a ligand bound octamer in m/z = 12,000–15,000. b, Mass spectrum of BT1762–BT1763 indicates that these two proteins form dimers and tetramers. The numbers in parentheses on the right are the theoretical masses. c, Analytical gel filtration chromatography of BT2261–64 (blue) and BT1762–BT1763 (green). For comparison, samples were run for soluble horse spleen ferritin (440 kDa; red) and for the membrane protein ammonium transporter Mep2 from Candida albicans (160 kDa; black). The following buffer was specifically for this experiment: 10 mM HEPES/100 mM NaCl/0.12% DM pH 7.5. Column: Superdex-200 Increase 10/300 GL. d, SEC–MALS analysis of BT1762–BT1763. Light scattering (LS) and differential refractive index (dRI) are plotted alongside the fitted total protein-detergent complex molecular weight (diamonds), and constituent protein (pluses) and detergent (crosses) molecular weights, across each peak. BT1762–BT1763 eluted as two species of 499 kDa (protein component 319 kDa, corresponding to a SusCD dimer) and 269 kDa (protein component 214 kDa). Chromatograms shown are from single experiments.
Extended Data Figure 5 Unbiased electron density for the bound ligand in BT2261–64.
Stereo views of simulated annealing omit maps using a starting temperature of 1000 K. a, 2Fo − Fc maps contoured at 1.5σ; carve, 2. b, Fo − Fc map contoured at 3.0σ; carve, 2. Selected residues contacting ligand are shown (yellow; BT2264/SusC, magenta, BT2263/SusD). Density for at least six amino acid side chains is present (denoted by an asterisk in the 2Fo − Fc map). c, Interaction table showing hydrogen-bond distances between the putative peptide ligand backbone and residues in BT2263 and BT2264.
Extended Data Figure 6 Molecular dynamics simulations for BT2261–64.
a, Plots of BT2264 (SusC) Cα r.m.s.d. versus simulation time for holo- and apo-complexes. b, c, Plots of BT2263 (SusD) Cα r.m.s.d. versus simulation time for holo and apo simulations, relative to the starting conformation (b) and after SusD superposition (c). d, e, Plots showing the number of hydrogen bonds between SusC and SusD versus simulation time (d) and between holo-SusCD and the modelled peptide (e). Simulations are numbered as follows: sim1–3, apo-BT2263–BT2264 (dimer); sim7–9, apo-BT2261–64 (tetramer); sim13, apo-(BT2261–64) × 2 (octamer); sim4–6, holo-BT2263–BT2264; sim10–12, holo-BT2261–64; sim14, holo-(BT2261–64) × 2. With the exception of those of the octamer (owing to its very large size), the simulations were repeated three times with different initial atomic velocities to allow sampling in order to obtain a measure of the possible spread in results.
Extended Data Figure 7 Dynamics of the bound peptide during molecular dynamics simulations.
a–c, Side views (left panels) and top views showing the bound peptide in the BT2263–BT2264 dimers (a), BT2261–64 tetramers (b) and the (BT2261–64) × 2 octamer (c). For clarity, only one final conformation for BT2264 (SusC) is shown together with the starting conformation of the peptide (green) and the final peptide conformations after 500 ns of simulation (red). For orientation purposes, the assigned N termini of the peptides are coloured blue.
Extended Data Figure 8 Molecular dynamics simulation root-mean-square-fluctuation analyses.
a, Cα root-mean-square-fluctuation values for SusC in holo- and apo-complexes with the conformational fluctuations of the hinge loop L7 highlighted separately. b, Cα root-mean-square-fluctuation values for SusD in apo and holo simulations. Simulations are numbered as in Extended Data Fig. 6: sim1–3, apo-BT2263–BT2264 (dimer); sim7–9, apo-BT2261–64 (tetramer); sim13, apo-(BT2261–64) × 2 (octamer); sim4–6, holo-BT2263–BT2264; sim10–12, holo-BT2261–64; sim14, holo-(BT2261–64) × 2.
Extended Data Figure 9 Structure of the BT2261–64 apo-octamer after 500 ns of molecular dynamics simulation (sim13), demonstrating the independent bin opening of the two SusCD hubs.
a, Views from the plane of the membrane rotated by 90°. b, View from the extracellular side. For clarity, the SusC and SusD subunits are shown in different colours (yellow and orange for SusC/BT2264, magenta and red for SusD/BT2263). BT2261 and BT2262 are shown in green and blue, respectively. c, Side view of the opened SusCD monomer highlighting the remaining interactions between SusC (yellow) and SusD (magenta) mediated by the SusC hinge loop L7 and loop L8.
Extended Data Figure 10 ITC analysis of levan binding for recombinant BT1762 SusD-like wild-type and mutants.
a, Titration curves from single experiments. Upper panels show raw injection heats of ligand (levan) into protein, lower panels show the integrated binding heats fit to a single set of sites binding model to determine Ka for all proteins except reduced wild type (10 mM TCEP), W85A and C298A mutants that display no binding. Levan stock solution was between 0.5–2% (w/v) and protein ranged from 50–60 μM. b, Levan affinity of recombinant BT1762 SusD-like wild type and mutant proteins determined by ITC. Ka values shown are averages and standard deviations from at least two independent titrations. Residue numbering is that of the mature protein (first residue Cys1).
Supplementary information
Supplementary Information
This file contains X-ray Crystallographic Tables showing data collection and refinement statistics for BT2261-64 (Supplementary Table 1), soluble proteins (Supplementary Table 2) and BT1762-63 (Supplementary Table 3). (PDF 50 kb)
Pedal bin opening of BT2263-64 (dimer)
Side-by-side movies showing 500 ns unbiased MD simulations of BT2263-64 in the presence (left; sim6) and absence (right; sim3) of the bound peptide ligand, shown as green spheres. BT2263 (SusD) is coloured magenta, BT2264 (SusC) yellow. The plug domain of SusC is coloured dark blue. (MOV 15918 kb)
Pedal bin opening of BT2261-64 (tetramer)
Side-by-side movies showing 500 ns unbiased MD simulations of BT2261-64 in the presence (left; sim11) and absence (right; sim8) of the bound peptide ligand, shown as green spheres. BT2263 (SusD) is coloured green, BT2264 (SusC) cyan. The small lipoproteins BT2261 and BT2262 are coloured grey and dark blue, respectively. (MOV 17469 kb)
Pedal bin opening of (BT2261-64)x 2 (octamer)
Side-by-side movies showing 500 ns unbiased MD simulations of 2 x (BT2261-64) in the presence (left; sim14) and absence (right; sim13) of the bound peptide ligand, shown as green spheres. BT2263 (SusD) is coloured green, BT2264 (SusC) cyan. The small lipoproteins BT2261 and BT2262 are coloured grey and dark blue, respectively. (MOV 19802 kb)
Rights and permissions
About this article
Cite this article
Glenwright, A., Pothula, K., Bhamidimarri, S. et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411 (2017). https://doi.org/10.1038/nature20828
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20828
This article is cited by
-
Particle-attached bacteria act as gatekeepers in the decomposition of complex phytoplankton polysaccharides
Microbiome (2024)
-
Successional action of Bacteroidota and Firmicutes in decomposing straw polymers in a paddy soil
Environmental Microbiome (2023)
-
A genetic system for Akkermansia muciniphila reveals a role for mucin foraging in gut colonization and host sterol biosynthesis gene expression
Nature Microbiology (2023)
-
Identifying glycan consumers in human gut microbiota samples using metabolic labeling coupled with fluorescence-activated cell sorting
Nature Communications (2023)
-
BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B12 uptake in gut Bacteroides
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.