Abstract
N6-methyladenosine (m6A) is the most common internal modification of eukaryotic messenger RNA (mRNA) and is decoded by YTH domain proteins1,2,3,4,5,6,7. The mammalian mRNA m6A methylosome is a complex of nuclear proteins that includes METTL3 (methyltransferase-like 3), METTL14, WTAP (Wilms tumour 1-associated protein) and KIAA1429. Drosophila has corresponding homologues named Ime4 and KAR4 (Inducer of meiosis 4 and Karyogamy protein 4), and Female-lethal (2)d (Fl(2)d) and Virilizer (Vir)8,9,10,11,12. In Drosophila, fl(2)d and vir are required for sex-dependent regulation of alternative splicing of the sex determination factor Sex lethal (Sxl)13. However, the functions of m6A in introns in the regulation of alternative splicing remain uncertain3. Here we show that m6A is absent in the mRNA of Drosophila lacking Ime4. In contrast to mouse and plant knockout models5,7,14, Drosophila Ime4-null mutants remain viable, though flightless, and show a sex bias towards maleness. This is because m6A is required for female-specific alternative splicing of Sxl, which determines female physiognomy, but also translationally represses male-specific lethal 2 (msl-2) to prevent dosage compensation in females. We further show that the m6A reader protein YT521-B decodes m6A in the sex-specifically spliced intron of Sxl, as its absence phenocopies Ime4 mutants. Loss of m6A also affects alternative splicing of additional genes, predominantly in the 5′ untranslated region, and has global effects on the expression of metabolic genes. The requirement of m6A and its reader YT521-B for female-specific Sxl alternative splicing reveals that this hitherto enigmatic mRNA modification constitutes an ancient and specific mechanism to adjust levels of gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Luo, S. & Tong, L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl Acad. Sci. USA 111, 13834–13839 (2014)
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012)
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012)
Perry, R. P. & Kelley, D. E. Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42 (1974)
Zhong, S. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008)
Schwartz, S. et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013)
Ke, S. et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 29, 2037–2053 (2015)
Liu, J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014)
Horiuchi, K. et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 288, 33292–33302 (2013)
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G. & Rottman, F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997)
Penalva, L. O. et al. The Drosophila fl(2)d gene, required for female-specific splicing of Sxl and tra pre-mRNAs, encodes a novel nuclear protein with a HQ-rich domain. Genetics 155, 129–139 (2000)
Niessen, M., Schneiter, R. & Nothiger, R. Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics 157, 679–688 (2001)
Schütt, C. & Nöthiger, R. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127, 667–677 (2000)
Geula, S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015)
Luo, G. Z. et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 5, 5630 (2014)
Xiao, W. et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016)
Hongay, C. F. & Orr-Weaver, T. L. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc. Natl Acad. Sci. USA 108, 14855–14860 (2011)
Bodi, Z., Bottley, A., Archer, N., May, S. T. & Fray, R. G. Yeast m6A methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS One 10, e0132090 (2015)
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015)
Meyer, K. D. et al. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 163, 999–1010 (2015)
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015)
Zaharieva, E., Haussmann, I. U., Bräuer, U. & Soller, M. Concentration and localization of co-expressed ELAV/Hu proteins control specificity of mRNA processing. Mol. Cell. Biol. 35, 3104–3115 (2015)
Salz, H. K. Sex, stem cells and tumors in the Drosophila ovary. Fly (Austin) 7, 3–7 (2013)
Bodi, Z., Button, J. D., Grierson, D. & Fray, R. G. Yeast targets for mRNA methylation. Nucleic Acids Res. 38, 5327–5335 (2010)
Hilfiker, A., Amrein, H., Dübendorfer, A., Schneiter, R. & Nöthiger, R. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 121, 4017–4026 (1995)
Starck, S. R. et al. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351, aad3867 (2016)
Church, C. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat. Genet. 42, 1086–1092 (2010)
Moindrot, B. et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Reports 12, 562–572 (2015)
Patil, D. P. et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016)
Haussmann, I. U., Hemani, Y., Wijesekera, T., Dauwalder, B. & Soller, M. Multiple pathways mediate the sex-peptide-regulated switch in female Drosophila reproductive behaviours. Proc. R. Soc. Lond. B 280, 20131938 (2013)
Haussmann, I. U., Li, M. & Soller, M. ELAV-mediated 3′-end processing of ewg transcripts is evolutionarily conserved despite sequence degeneration of the ELAV-binding site. Genetics 189, 97–107 (2011)
Yan, D. & Perrimon, N. spenito is required for sex determination in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 112, 11606–11611 (2015)
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods 25, 402–408 (2001)
Haussmann, I. U., White, K. & Soller, M. Erect wing regulates synaptic growth in Drosophila by integration of multiple signaling pathways. Genome Biol. 9, R73 (2008)
Soller, M. & White, K. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 17, 2526–2538 (2003)
Harper, J. E., Miceli, S. M., Roberts, R. J. & Manley, J. L. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic Acids Res. 18, 5735–5741 (1990)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012)
Sturgill, D. et al. Design of RNA splicing analysis null models for post hoc filtering of Drosophila head RNA-seq data with the splicing analysis kit (Spanki). BMC Bioinformatics 14, 320 (2013)
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011)
Soller, M., Bownes, M. & Kubli, E. Control of oocyte maturation in sexually mature Drosophila females. Dev. Biol. 208, 337–351 (1999)
Soller, M. & White, K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol. Cell. Biol. 25, 7580–7591 (2005)
Acknowledgements
We thank J. Horabin, N. Perrimon and the Bloomington, Harvard and Kyoto stock centres for fly lines, BacPAc for DNA clones, E. Zaharieva and M. L. Li for help with imaging, W. Arlt and R. Michell for comments on the manuscript, and J.-Y. Roignant for communication of results before publication. We acknowledge funding from the BBSRC (BB/M008606/1) to R.F.
Author information
Authors and Affiliations
Contributions
I.U.H. and M.S. performed biochemistry, cell biology and genetic experiments, E.S.M. stained chromosomes, and Z.B., N.A. and R.F. performed biochemistry experiments. N.M. analysed sequencing data. I.U.H., R.F. and M.S. conceived the project and wrote the manuscript with help from N.M. and Z.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 m6A levels in unfertilized eggs.
a, b, Thin-layer chromatography from maternal total RNA (a) and mRNA (b) present in unfertilized eggs. The arrow indicates m6A.
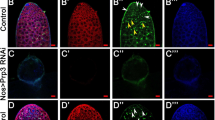
Extended Data Figure 2 Ime4 supports Sxl in directing germline differentiation.
a–c, Representative ovarioles of wild-type (a), Ime4null/Ime4null (b) and Sxl/+;Ime4null/+ females (c), and a tumerous ovary of a Sxl/+;Ime4null/+ female (d). The tumorous ovary consisting mostly of undifferentiated germ cells in d is indicated with a bracket and the oviduct with an asterisk. Scale bar, 100 μm (applies to all panels).
Extended Data Figure 3 Ime4 is required for female-specific splicing of Sxl, tra and msl-2.
a–c, RT–PCR of Sxl (a), tra (b) and msl-2 (c) sex-specific splicing in wild-type males and females, and Ime4null males and females. 100-bp markers are shown on the left. AS, alternative splicing.
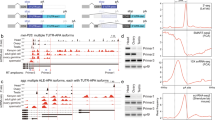
Extended Data Figure 4 Alternative splicing of sex-determination genes and differential expression of X-linked genes in Ime4null females.
a–c, Sashimi plot depicting Tophat-mapped RNA sequencing reads and exon junction reads below the annotated gene model for sex-specific alternative splicing of tra, fru and dsx. The thickness of lines connecting splice junctions corresponds to the number of junction reads also shown. ss, splice site. d, Significantly (P < 0.05, q < 0.166853) differentially expressed gene expression values expressed as reads per kb of transcript per million mapped reads (RPKM) were log[x + 1]-transformed and Spearman r correlation values determined for X-linked and autosomal genes in wild-type and Ime4null Drosophila. e, The proportion of autosomal and X-linked genes that were significantly either up- or downregulated in Ime4null as compared to wild-type Drosophila were statistically compared using χ2 with Yates’ continuity correction. GraphPad Prism was used for statistical comparisons. Similar results as for the single-read RNA-seq experiment were obtained for the paired-end RNA sequencing experiment.
Extended Data Figure 5 m6A methylation sites map to the vicinity of Sxl binding sites.
a, Schematic of the Sxl alternatively spliced intron around the male-specific exon depicting substrate RNAs used for in vitro m6A methylation. Solid lines depict fragments containing m6A methylation and dashed lines indicate fragments where m6A was absent. b, c, 1D-TLC of in vitro methylated [32P]-ATP-labelled substrate RNAs shown in a. Markers are in vitro transcripts in the absence (M1) or presence (M2) of m6A 32P-labelled after RNase T1 digestion. The right panels in b and c show an overexposure of the same thin-layer chromatography.
Extended Data Figure 6 RT–PCR validation of differential alternative splicing in Ime4null flies.
a–f, Sashimi plots depicting Tophat-mapped RNA sequencing reads and exon junction reads below the annotated gene model of indicated genes on the left, and RT–PCR of alternative splicing shown on the right using primers depicted on top. The thickness of lines connecting splice junctions corresponds to the number of junction reads also shown.
Extended Data Figure 7 Ime4 affects alternative splicing predominantly in 5′ UTRs in genes with a higher than average number of upstream start codons.
a, b, Classification of differential alternative splicing in Ime4null according to splicing event (a) and location of the event in the mRNA (b). c, Quantification of upstream start codons (AUGs) in all annotated 5′ UTRs (white) or in alternative isoforms differentially spliced between wild-type and Ime4null insects. All Drosophila UTRs were accessed in fasta format from Flybase (version r6.07), (ftp://ftp.flybase.net/genomes/Drosophila_melanogaster/current/fasta/). An R script was used to count the number of ATG sequences in all Drosophila 5′ UTRs and from the genes identified by the Spanki analysis comprising 638 5′ UTRs. A t-test was then used to statistically compare the number of ATGs present in the 638 5′ UTRs of the differentially spliced genes as compared to all 29,822 Drosophila 5′ UTRs. d, e, Classification of differentially alternatively spliced genes in Ime4null according to expression pattern (d) or function (e).
Extended Data Figure 8 Drosophila S2 cells are male.
RT–PCR of Sxl alternative splicing in females, males and S2 cells. 100-bp markers are shown on the left.
Extended Data Figure 9 Preferential binding of the YTH domain of YT521-B to m6A-containing RNA.
a, Coomassie-stained gel depicting the recombinant YTH domain (amino acids 207–423) of YT521-B. b, c, Electrophoretic mobility shift assay of YTH domain binding to Sxl RNA fragment C with or without m6A (50% of adenosine in the transcript methylated) and quantification of RNA bound to the YTH domain shown as mean ± s.e.m. (n = 3). Note that the YTH domain does not form a stable complex with RNA (asterisk) and that this complex falls apart during the run or forms aggregates in the well. d, UV cross-linking of the YTH domain to Sxl RNA fragment C at 0.25 μM, 1 μM, 4 μM and 16 μM (lanes 1–4).
Extended Data Figure 10 YT521-B co-localizes to sites of transcription.
a–d, Polytene chromosomes from salivary glands expressing YT521-B::HA stained with anti-Pol II (red, b), anti-HA (green, c) and DNA (DAPI, blue, d), or merged (yellow, a). Scale bars, 5 μm.
Supplementary information
Supplementary Information
This file contains graphical source data, uncropped gels, Western blots and 1D TLCs. (PDF 960 kb)
Supplementary Table 1
This file contains the alternative splicing analysis. (XLS 168 kb)
Supplementary Table 2
This file contains the differential gene expression analysis. (XLS 154 kb)
Rights and permissions
About this article
Cite this article
Haussmann, I., Bodi, Z., Sanchez-Moran, E. et al. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304 (2016). https://doi.org/10.1038/nature20577
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20577
This article is cited by
-
The role of the methyltransferase METTL3 in prostate cancer: a potential therapeutic target
BMC Cancer (2024)
-
C2-methyladenosine in tRNA promotes protein translation by facilitating the decoding of tandem m2A-tRNA-dependent codons
Nature Communications (2024)
-
New horizons for the role of RNA N6-methyladenosine modification in hepatocellular carcinoma
Acta Pharmacologica Sinica (2024)
-
N6-methyladenosine-induced miR-182-5p promotes multiple myeloma tumorigenesis by regulating CAMK2N1
Molecular and Cellular Biochemistry (2024)
-
The role of m6A epigenetic modifications in tumor coding and non-coding RNA processing
Cell Communication and Signaling (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.