Abstract
The type III secretion (T3S) injectisome is a specialized protein nanomachine that is critical for the pathogenicity of many Gram-negative bacteria, including purveyors of plague, typhoid fever, whooping cough, sexually transmitted infections and major nosocomial infections. This syringe-shaped 3.5-MDa macromolecular assembly spans both bacterial membranes and that of the infected host cell. The internal channel formed by the injectisome allows for the direct delivery of partially unfolded virulence effectors into the host cytoplasm1. The structural foundation of the injectisome is the basal body, a molecular lock-nut structure composed predominantly of three proteins that form highly oligomerized concentric rings spanning the inner and outer membranes2,3,4,5. Here we present the structure of the prototypical Salmonella enterica serovar Typhimurium pathogenicity island 1 basal body, determined using single-particle cryo-electron microscopy, with the inner-membrane-ring and outer-membrane-ring oligomers defined at 4.3 Å and 3.6 Å resolution, respectively. This work presents the first, to our knowledge, high-resolution structural characterization of the major components of the basal body in the assembled state, including that of the widespread class of outer-membrane portals known as secretins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Worrall, L. J., Lameignere, E. & Strynadka, N. C. Structural overview of the bacterial injectisome. Curr. Opin. Microbiol. 14, 3–8 (2011)
Bergeron, J. R. et al. A refined model of the prototypical Salmonella SPI-1 T3SS basal body reveals the molecular basis for its assembly. PLoS Pathog. 9, e1003307 (2013)
Bergeron, J. R. et al. The modular structure of the inner-membrane ring component PrgK facilitates assembly of the type III secretion system basal body. Structure 23, 161–172 (2015)
Spreter, T. et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat. Struct. Mol. Biol. 16, 468–476 (2009)
Yip, C. K. et al. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435, 702–707 (2005)
Korotkov, K. V., Gonen, T. & Hol, W. G. Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36, 433–443 (2011)
Schraidt, O. et al. Topology and organization of the Salmonella Typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824 (2010)
Schraidt, O. & Marlovits, T. C. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195 (2011)
Sanowar, S. et al. Interactions of the transmembrane polymeric rings of the Salmonella enterica serovar Typhimurium type III secretion system. MBio 1, (2010)
Korotkov, K. V., Pardon, E., Steyaert, J. & Hol, W. G. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265 (2009)
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007)
Linderoth, N. A., Model, P. & Russel, M. Essential role of a sodium dodecyl sulfate-resistant protein IV multimer in assembly-export of filamentous phage. J. Bacteriol. 178, 1962–1970 (1996)
Hardie, K. R., Lory, S. & Pugsley, A. P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15, 978–988 (1996)
Guilvout, I. et al. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J. Mol. Biol. 382, 13–23 (2008)
Guilvout, I., Nickerson, N. N., Chami, M. & Pugsley, A. P. Multimerization-defective variants of dodecameric secretin PulD. Res. Microbiol. 162, 180–190 (2011)
Koo, J., Burrows, L. L. & Howell, P. L. Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328, 1–12 (2012)
Okon, M. et al. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure 16, 1544–1554 (2008)
Gu, S., Rehman, S., Wang, X., Shevchik, V. E. & Pickersgill, R. W. Structural and functional insights into the pilotin-secretin complex of the type II secretion system. PLoS Pathog. 8, e1002531 (2012)
Huysmans, G. H., Guilvout, I., Chami, M., Nickerson, N. N. & Pugsley, A. P. Lipids assist the membrane insertion of a BAM-independent outer membrane protein. Sci. Rep. 5, 15068 (2015)
Hu, B. et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl Acad. Sci. USA 112, 1047–1052 (2015)
Hoang, H. H. et al. Outer membrane targeting of Pseudomonas aeruginosa proteins shows variable dependence on the components of Bam and Lol machineries. MBio 2, (2011)
Dunstan, R. A. et al. Assembly of the secretion pores GspD, Wza and CsgG into bacterial outer membranes does not require the Omp85 proteins BamA or TamA. Mol. Microbiol. 97, 616–629 (2015)
Guilvout, I. et al. Independent domain assembly in a trapped folding intermediate of multimeric outer membrane secretins. Structure 22, 582–589 (2014)
Huysmans, G. H., Guilvout, I. & Pugsley, A. P. Sequential steps in the assembly of the multimeric outer membrane secretin PulD. J. Biol. Chem. 288, 30700–30707 (2013)
Nans, A., Kudryashev, M., Saibil, H. R. & Hayward, R. D. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat. Commun. 6, 10114 (2015)
Disconzi, E. et al. Bacterial secretins form constitutively open pores akin to general porins. J. Bacteriol. 196, 121–128 (2014)
Papahadjopoulos, D., Moscarello, M., Eylar, E. H. & Isac, T. Effects of proteins on thermotropic phase transitions of phospholipid membranes. Biochim. Biophys. Acta 401, 317–335 (1975)
Burghout, P. et al. Structure and electrophysiological properties of the YscC secretin from the type III secretion system of Yersinia enterocolitica. J. Bacteriol. 186, 4645–4654 (2004)
Spagnuolo, J. et al. Identification of the gate regions in the primary structure of the secretin pIV. Mol. Microbiol. 76, 133–150 (2010)
Marlovits, T. C. et al. Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 (2004)
Reichow, S. L., Korotkov, K. V., Hol, W. G. & Gonen, T. Structure of the cholera toxin secretion channel in its closed state. Nat. Struct. Mol. Biol. 17, 1226–1232 (2010)
Loquet, A. et al. Atomic model of the type III secretion system needle. Nature 486, 276–279 (2012)
Chang, Y. W. et al. Architecture of the type IVa pilus machine. Science 351, aad2001 (2016)
Kimbrough, T. G. & Miller, S. I. Contribution of Salmonella Typhimurium type III secretion components to needle complex formation. Proc. Natl Acad. Sci. USA 97, 11008–11013 (2000)
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005)
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015)
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
DiMaio, F. et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015)
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010)
Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001)
Chan, Q. W., Howes, C. G. & Foster, L. J. Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5, 2252–2262 (2006)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008)
Edwards, R. A., Keller, L. H. & Schifferli, D. M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 (1998)
Daefler, S. & Russel, M. The Salmonella Typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol. Microbiol. 28, 1367–1380 (1998)
Hodgkinson, J. L. et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat. Struct. Mol. Biol. 16, 477–485 (2009)
Kowal, J. et al. Structure of the dodecameric Yersinia enterocolitica secretin YscC and its trypsin-resistant core. Structure 21, 2152–2161 (2013)
Tosi, T. et al. Structural similarity of secretins from type II and type III secretion systems. Structure 22, 1348–1355 (2014)
Vizcaíno, J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013)
Acknowledgements
We thank A. Cheung for assistance with the expression trials of the PrgH130–392 GFP-fused basal body, C. Lizak for advice on GFP variants and membrane-protein purification and C. Yip and J. Rubenstein for advice on negative-stain TEM techniques. We thank UBC Bioimaging for access to TEM infrastructure. We thank K.-M. Moon and J. Rogalski at the Michael Smith Labs Proteomics Core Facility for assistance with LC–MS/MS. We thank F. Rosell at the LMB Spectroscopy and Kinetics Hub for assistance with circular dichroism. We thank S. Miller for providing the S. Typhimurium deletion strains and plasmids, as well as the InvG antibody. This work was funded by scholarships to L.W. and J.B. from the Canadian Institutes of Health Research (CIHR) and Michael Smith Foundation of Health Research, respectively, and operating grants from CIHR to N.C.J.S. and B.B.F., and the Howard Hughes International Senior Scholar program to N.C.J.S. N.C.J.S. is a Tier I Canada Research Chair in Antibiotic Discovery.
Author information
Authors and Affiliations
Contributions
L.J.W. performed all model building, refinement, structural analysis and modelling experiments. C.H. performed single particle cryo-EM grid preparation, data collection and map generation with input from R.K.H. and Z.Y. M.V. performed all cloning, basal body and secretin sample preparations used in the structure solution with input from L.J.W. and J.R.C.B., building on work initiated by T.S. L.J.W. and J.R.C.B. carried out negative-stain TEM analysis on basal body and secretin preps for quality control. L.J.W. and M.V. designed and made basal body mutants for secretion assays carried out by W.D. W.D. also generated/validated the invG deletion mutant. D.D.M. performed experiments probing isolated pilotin InvH and InvG S domain interactions. L.J.W. and N.C.J.S. principally wrote the manuscript with input from all.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Beeby, A. Blocker, J. Rubinstein and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
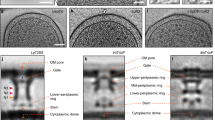
Extended Data Figure 1 3D reconstructions of the basal body complex.
a, Representative micrograph of the basal body complex (a total of 2,515 were recorded). b, Selected reference-free 2D class averages. c, d, The side view (c) and the bottom view (d) of the C1 (no symmetry imposed) reconstructed map of the basal body complex. e, f, The side view (e) and the bottom view (f) of the C24 (24-fold symmetry imposed) reconstructed map of the basal body complex. g, h, FSC of the C1 reconstruction (g) and the C24 reconstruction (h) calculated in Frealign using unmasked maps. i, Local resolution estimations of the C1 map. j, The C24 map from ResMap. Arrows in c and e indicate location of slice. k, Representative density for the inner-membrane rings (4.3 Å resolution).
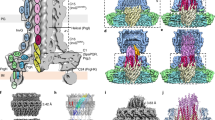
Extended Data Figure 2 PrgH130–392 basal body composition and oligomerization of PrgH and PrgK.
a, Proteins from the purified PrgH130–392 basal body identified by LC–MS/MS. InvG, PrgH, PrgK and SpaP were detected with elevated intensity. InvA and SpaQ peptides were also detected with decreased abundance. Some cytoplasmic export apparatus proteins and effectors were detected at very low levels, indicating they can still interact with the basal body complex. Only T3SS proteins are shown. *MW of PrgH130–392. b, Schematic of PrgH and PrgK domain topology and position, as observed in our basal body structure. The PrgH cytoplasmic D1 domain is absent in the PrgH130–392 mutant and its location with respect to the basal body is unclear. c, Structure of PrgH171–364 (green) and PrgK20–203 (orange). Previously unresolved PrgK loops now observed in the assembled state are labelled and shown as sticks. d, Role of the PrgK D1–D2 linker in PrgK oligomerization. Residues 84–90 form a helix, with Phe89 (shown as spheres) inserting between neighbouring D2 domains. e, The D2 transmembrane loop contributes to oligomerization interacting with both the PrgK D1 domain and neighbouring PrgH protomers (green/yellow). Leu195 is the equivalent termination position in EPEC EscJ and the local environment of Lys203 was previously supported by chemical cross-linking9.
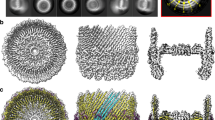
Extended Data Figure 3 3D reconstruction of the isolated secretin.
a, Representative micrograph of the isolated secretin (total 2,685 recorded). b, Selected reference-free 2D class averages. c, d, The bottom view (c) and side view (d) of the C1 (no symmetry imposed) reconstructed map of the isolated secretin. e, FSC curve of the C1 reconstruction using gold-standard refinement with soft-masking-effect correction. f, g, Bottom view (f) and, side view (g) of the C15 (15-fold symmetry imposed) reconstructed map of the isolated secretin. h, FSC curve of the C15 reconstruction using gold-standard refinement with soft-masking-effect correction. i, Representative density for the isolated secretin (3.6 Å resolution).
Extended Data Figure 4 Secondary structure topology of the InvG secretin.
Secondary structure topology for InvG172–557. β-strands of the secretin domain are numbered, with 1, 3a/3b, 8 and 9 forming the outer β-barrel; 4–7 forming the inner β-barrel; and 1, 2 and 3a forming the lip of the β-barrel. Strand 3 is broken into 3a and 3b by the conserved residue Pro371.
Extended Data Figure 5 Sequence conservation of secretins and structure-based mapping of previously characterized PulD oligomerization mutants.
a, InvG172–557 sequence conservation (see Methods), coloured from magenta (highest) to cyan (lowest) for T3SS homologues. b, c, Close-up views of boxed regions in a. Pro371 and Gly471 (see f), highly conserved in all secretins (see Extended Data Fig. 6a), are labelled in c. d, PulD multimerization mutants mapped onto the InvG structure (shown as spheres). Coloured according to Fig. 2. Residues are labelled with letters and correspondingly annotated in e (N3 domain alignment; a–b) or Extended Data Fig. 6a (secretin domain alignment; c–t). e, Alignment of N3 domain (InvG180–300) from T3SS (green) and non-T3SS (orange) secretin homologues (see Fig. 3d). Conserved regions are lettered red and boxed. Invariant residues are boxed in solid red. Secondary structural elements observed here are annotated and numbered, consistent with other RBMs. PulD multimerization and pIV permeability mutants mapping to this domain are indicated by letters or numbers, respectively, and shown in d (for PulD) or Extended Data Fig. 10b (for pIV). f, InvG complementation assay for N3 domain ring-interface mutants (Leu293Arg, Leu293Ala; labelled in d) and conserved secretin domain β sandwich mutants (Pro371Leu, Gly471Ala; also labelled in c and Extended Data Fig. 6a). Lysate InvG protein levels indicated below using anti-InvG antibody.
Extended Data Figure 6 Sequence alignment for T3SS and non-T3SS secretin domains.
a, Alignment of secretin domain (InvG302–519) from T3SS (green) and non-T3SS (orange) secretin homologues (see Fig. 3d). Anti-parallel strands of the outer and inner β-sheets coloured cyan and green respectively with loop regions (where most indels are located) denoted by pattern of diagonal lines. Strands numbered according to Extended Data Fig. 4. Conserved residues Pro371 and Gly471 (Extended Data Fig. 5f) are marked with asterisks. PulD multimerization and pIV permeability mutants mapping to this domain are indicated by letters or numbers respectively and shown in Extended Data Fig. 5d (PulD) or Extended Data Fig. 10b (pIV). b, Corresponding structural elements. The residues that make up the putative membrane interacting region (gold) are boxed in both the structure and the sequence alignment.
Extended Data Figure 7 Secretin oligomerization and stoichiometry.
a, Secretin domain interface between neighbouring monomers (denoted as i and i + 1). To differentiate adjacent neighbours and their extensive interfaces, the i ribbon is coloured as in Fig. 2 with the residues forming the oligomeric interfaces shown as sticks. The i + 1 ribbon is coloured grey with the residues at the oligomeric interface shown as transparent Van der Waal spheres and coloured according to domain feature. b, c, Role of N3 (cobalt blue) and S (red) domains in secretin multimerization. The i ribbon is coloured as in Fig. 2 with N3 and S domain interface residues shown as sticks, neighbouring monomers (i + 1 and i + 2) coloured grey and white, respectively (and also delineated by dashed line), with the residues interacting with N3 or S domains from monomer i displayed as spheres and coloured as in Fig. 2. The N3 domain of monomer i (blue sticks) interfaces with both the N3 domain (blue spheres) and underside of the inner β sheet (green spheres) of monomer i + 1. The S domain of monomer i (red sticks) forms an extensive stapled interface with the outer β-sheet of i + 1 and i + 2 monomers (cyan spheres) and with the N-terminal S domain helix of monomer i + 1 (red spheres). d, e, Superimposition of available secretin electron microscopy maps and table showing EMD accession number, resolution, stoichiometry (n) and molecular weight (MW) of the secretin domain monomer. All secretins have the same general architecture, with GspD having an elongated upper lip and enclosed upper chamber as a result of the loop 1 insertion following strand 2 (Extended Data Fig. 6a). MxiD (S. flexneri T3SS), YscC (Y. enterocolitica T3SS), PscC (P. aeruginosa T3SS), GspD (V. cholerae T2SS), PulD (K. oxytoca T2SS).
Extended Data Figure 8 Interaction of the secretin S domain with the pilotin.
a, InvG S domain (red) superposed with the MxiM/MxiD pilotin-bound secretin peptide (yellow) from S. flexneri. The electronegative MxiD Glu555 and Asp556 superpose with InvG Asp543 and Asp544. InvG Asp544 forms a salt bridge with Lys315, which is also conserved between MxiD and InvG (shown as sticks). Ordering of this helical peptide of the secretin S domain has been shown to occur upon binding to the pilotin. b, Superimposition of pilotin MxiM (green surface) onto InvG, based on binding to a common secretin peptide as in a, showing the S domain orientation is permissive of an assembled interaction with pilotin in the oligomerized form. Putative linker (solid line) and membrane inserted lipidation shown. OM, outer membrane. c, Far-UV circular dichroism spectra (from an average of four scans) for InvG520–562 (blue line), InvH27–147 (green line) and the complex (red line) showing an approximately 20% increase in ellipticity at 222 nM as compared to the calculated combined spectra (red dashed line), indicative of increased helical content in the complex and consistent with a disorder–order transition in the InvG S domain. d, e, ITC analysis for the interaction between InvH27–147 and InvG520–562 and InvG520–562(K548A/W549A/V552A) respectively. A representative run is shown (from four runs) and Kd and N (stoichiometry) are reported as mean (standard deviation) from four runs. f, Generalized schematic of pilotin domains. N-terminal type II signal sequence followed by a lipobox lipidation signal with conserved cysteine connected to a structurally diverse globular C-terminal secretin binding domain via a variable length linker. g, InvG complementation assay for S domain deletion mutants and C-terminal helix triple mutant. Lysate InvG protein levels indicated below using anti-InvG antibody.
Extended Data Figure 9 InvG membrane localization and proposed pathway for pore assembly and membrane insertion.
a, Distribution of wild-type (WT) InvG and mutants in whole-cell lysate, soluble and membrane fractions. Ability to form SDS-resistant oligomer assessed by running both boiled (+) and unboiled (−) samples. The AHL mutant InvG(F486A/L487A/L490A/L492A/I493A/L496A/F497A) and transmembrane β-strand mutant InvG(V337G/L339G) (mutants that abrogate and reduce secretion, respectively; see Fig. 3e) both localize to the membrane (the AHL mutant to a lesser degree); however, their ability to form SDS-resistant oligomers is substantially affected and protease sensitivity is evident for the AHL mutant—both observations are consistent with aberrant membrane association, insertion and final stabilized assembly24. b–d, Schematic of the proposed secretin assembly and membrane insertion pathway. The secretin monomer is localized to the outer membrane in a Lol-dependent manner, most likely in a complex with the pilotin, where we propose initial membrane association is mediated by the conserved amphipathic loop (AHL; gold). Oligomerization to a membrane associated pre-pore intermediate follows which, based on our structure and earlier negative stain electron microscopy of this form in PulD (c; figure reproduced from ref. 23), would encompass a folded secretin core and the peripheral N3 and S domains with the extensive interfaces between them (see Extended Data Fig. 7a–c) stabilizing this pre-pore form. Subsequent folding of the remainder of the secretin β-domain (disordered in the pre-pore image) to create the upper β-barrel lip (gold) leads to the final, BAM-independent, membrane insertion and creation of the membrane-spanning secretin pore. Outer-membrane curvature observed in situ in several T3SSs is illustrated by overlaying the secretin structure with the in situ tomography of the Shigella injectisome (d; figure reproduced from ref. 20) also showing presence of continuous density, proposed to be the pilotin (circled), connecting the inner leaflet of the outer membrane and the region corresponding to the S domain in our InvG structure.
Extended Data Figure 10 The secretin periplasmic gate.
a, Superimposition of the structure of InvG (coloured according to Fig. 2), basal body map (cyan), the approximately 10 Å needle-complex map (EMD-2481; grey) and the approximately 20 Å needle-complex map (EMD-1100; pink). Our basal body has dimensions with greater similarity to EMD-1100. Substrate passage through the secretin would require N3 domain reorientation and subsequent opening of periplasmic gate indicated with black arrows. Differences in the regions corresponding to the upper secretin β-sandwich in the needle-complex maps (circled) suggest a conformation of the open gate packed against the outer β-sheet. b, InvG structure, showing the periplasmic gate and N3 domain residues implicated in gating. InvG mutants with a secretion-deficient phenotype (shown in c) are shown as balls and sticks and the location of the N3–secretin interface-localized phage secretin pIV permeability mutants (additional pIV permeability mutants are mapped throughout the inner β-sheet) are shown as spheres (labelled 1–4 and correspondingly annotated in Extended Data Fig. 5e (N3 domain alignment; 1–2) or Extended Data Fig. 6a (secretin domain alignment; 3–4)). c, InvG complementation assay for periplasmic-gate and N3-hairpin mutants. Lysate InvG protein levels are indicated below using anti-InvG antibodies. d, Electrostatic surface of the InvG pore structure, including the Rosetta-modelled N0 and N1 domains, as viewed from the periplasmic face. e, Electrostatic surface (left) of the Salmonella SPI-1 needle (PDB 2LPZ) and corresponding ribbon representation (right). A single PrgI monomer is coloured magenta. f, Superimposition of Salmonella SPI-1 needle (purple) onto the InvG172–557 pore structure (blue), as viewed from the extracellular face.
Supplementary information
Supplementary Information
This file contains original SDS-PAGE gels and anti-InvG western blots used to generate Fig. 3e (a, b), Extended Data Fig. 5f (a, b), Extended Data Fig. 8g (a, b), Extended Data Fig. 10c (c, d) and Extended Data Fig. 9a (e). (PDF 4478 kb)
Rights and permissions
About this article
Cite this article
Worrall, L., Hong, C., Vuckovic, M. et al. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540, 597–601 (2016). https://doi.org/10.1038/nature20576
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20576
This article is cited by
-
Surveying membrane landscapes: a new look at the bacterial cell surface
Nature Reviews Microbiology (2023)
-
Structure of a type IV secretion system core complex encoded by multi-drug resistance F plasmids
Nature Communications (2022)
-
Type three secretion system in Salmonella Typhimurium: the key to infection
Genes & Genomics (2020)
-
Architecture of type VI secretion system membrane core complex
Cell Research (2019)
-
Intermembrane crosstalk drives inner-membrane protein organization in Escherichia coli
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.