Abstract
Sleep is conserved from invertebrates to vertebrates, and is tightly regulated in a homeostatic manner. The molecular and cellular mechanisms that determine the amount of rapid eye movement sleep (REMS) and non-REMS (NREMS) remain unknown. Here we identify two dominant mutations that affect sleep and wakefulness by using an electroencephalogram/electromyogram-based screen of randomly mutagenized mice. A splicing mutation in the Sik3 protein kinase gene causes a profound decrease in total wake time, owing to an increase in inherent sleep need. Sleep deprivation affects phosphorylation of regulatory sites on the kinase, suggesting a role for SIK3 in the homeostatic regulation of sleep amount. Sik3 orthologues also regulate sleep in fruitflies and roundworms. A missense, gain-of-function mutation in the sodium leak channel NALCN reduces the total amount and episode duration of REMS, apparently by increasing the excitability of REMS-inhibiting neurons. Our results substantiate the use of a forward-genetics approach for studying sleep behaviours in mice, and demonstrate the role of SIK3 and NALCN in regulating the amount of NREMS and REMS, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Cirelli, C. et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092 (2005)
Koh, K. et al. Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372–376 (2008)
Raizen, D. M. et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572 (2008)
Daan, S., Beersma, D. G. & Borbély, A. A. Timing of human sleep: recovery process gated by a circadian pacemaker. Am. J. Physiol. 246, R161–R183 (1984)
Franken, P., Chollet, D. & Tafti, M. The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 21, 2610–2621 (2001)
Suzuki, A., Sinton, C. M., Greene, R. W. & Yanagisawa, M. Behavioral and biochemical dissociation of arousal and homeostatic sleep need influenced by prior wakeful experience in mice. Proc. Natl Acad. Sci. USA 110, 10288–10293 (2013)
Lu, J., Sherman, D., Devor, M. & Saper, C. B. A putative flip-flop switch for control of REM sleep. Nature 441, 589–594 (2006)
Saper, C. B., Scammell, T. E. & Lu, J. Hypothalamic regulation of sleep and circadian rhythms. Nature 437, 1257–1263 (2005)
Luppi, P. H. et al. The neuronal network responsible for paradoxical sleep and its dysfunctions causing narcolepsy and rapid eye movement (REM) behavior disorder. Sleep Med. Rev. 15, 153–163 (2011)
Xu, M. et al. Basal forebrain circuit for sleep-wake control. Nat. Neurosci. 18, 1641–1647 (2015)
Adamantidis, A. R., Zhang, F., Aravanis, A. M., Deisseroth, K. & de Lecea, L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450, 420–424 (2007)
Herrera, C. G. et al. Hypothalamic feedforward inhibition of thalamocortical network controls arousal and consciousness. Nat. Neurosci. 19 290–298 (2016)
Carter, M. E. et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 13, 1526–1533 (2010)
Weber, F. et al. Control of REM sleep by ventral medulla GABAergic neurons. Nature 526, 435–438 (2015)
Hayashi, Y. et al. Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science 350, 957–961 (2015)
Takahashi, J. S., Shimomura, K. & Kumar, V. Searching for genes underlying behavior: lessons from circadian rhythms. Science 322, 909–912 (2008)
Citri, Y. et al. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326, 42–47 (1987)
King, D. P. et al. Positional cloning of the mouse circadian Clock gene. Cell 89, 641–653 (1997)
Allada, R., Emery, P., Takahashi, J. S. & Rosbash, M. Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci. 24, 1091–1119 (2001)
Kumar, V. et al. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342, 1508–1512 (2013)
Takemori, H. & Okamoto, M. Regulation of CREB-mediated gene expression by salt inducible kinase. J. Steroid Biochem. Mol. Biol. 108, 287–291 (2008)
Vyazovskiy, V. V. et al. Local sleep in awake rats. Nature 472, 443–447 (2011)
Katoh, Y. et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 273, 2730–2748 (2006)
Berggreen, C., Henriksson, E., Jones, H. A., Morrice, N. & Göransson, O. cAMP-elevation mediated by β-adrenergic stimulation inhibits salt-inducible kinase (SIK) 3 activity in adipocytes. Cell. Signal. 24, 1863–1871 (2012)
Flourakis, M. et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162, 836–848 (2015)
Ren, D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron 72, 899–911 (2011)
Lu, B. et al. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature 457, 741–744 (2009)
Lu, B. et al. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron 68, 488–499 (2010)
Crochet, S., Onoe, H. & Sakai, K. A potent non-monoaminergic paradoxical sleep inhibitory system: a reverse microdialysis and single-unit recording study. Eur. J. Neurosci. 24, 1404–1412 (2006)
Sapin, E. et al. Localization of the brainstem GABAergic neurons controlling paradoxical (REM) sleep. PLoS One 4, e4272 (2009)
Krishnan, K. S. & Nash, H. A. A genetic study of the anesthetic response: mutants of Drosophila melanogaster altered in sensitivity to halothane. Proc. Natl Acad. Sci. USA 87, 8632–8636 (1990)
Lear, B. C. et al. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48, 965–976 (2005)
Joiner, W. J. et al. Genetic and anatomical basis of the barrier separating wakefulness and anesthetic-induced unresponsiveness. PLoS Genet. 9, e1003605 (2013)
Weber, F. & Dan, Y. Circuit-based interrogation of sleep control. Nature 538, 51–59 (2016)
Funato, H. et al. Loss of Goosecoid-like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proc. Natl Acad. Sci. USA 107, 18155–18160 (2010)
Franken, P., Malafosse, A. & Tafti, M. Genetic variation in EEG activity during sleep in inbred mice. Am. J. Physiol. 275, R1127–R1137 (1998)
Funato, H., Saito-Nakazato, Y. & Takahashi, H. Axonal growth from the habenular nucleus along the neuromere boundary region of the diencephalon is regulated by semaphorin 3F and netrin-1. Mol. Cell. Neurosci. 16, 206–220 (2000)
Mashiko, D. et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3, 3355 (2013)
Lu, B. et al. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129, 371–383 (2007)
Wang, B. et al. A hormone-dependent module regulating energy balance. Cell 145, 596–606 (2011)
Kume, K., Kume, S., Park, S. K., Hirsh, J. & Jackson, F. R. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25, 7377–7384 (2005)
Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl Acad. Sci. USA 98, 12596–12601 (2001)
Lanjuin, A. & Sengupta, P. Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33, 369–381 (2002)
Singh, K. et al. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr. Biol. 21, 825–834 (2011)
Acknowledgements
We thank all Yanagisawa/Funato laboratory and WPI-IIIS members, especially T. Motoike, A. Matsui, Y. Goto, M. Takahashi and K. Taniguchi for technical assistance and their participation in the early stage of this project, M. Lazarus, R. W. Greene and K. E. Vogt for discussion and comments on this manuscript. J.S.T. is an investigator and M.Y. is a former investigator of the Howard Hughes Medical Institute. This work was supported by the World Premier International Research Center Initiative from MEXT to M.Y., JSPS KAKENHI (grant number 26220207 to M.Y., H.F., T.K.; 16K15187 to H.F.; 26507003 to C.M., H.F.; 15K18966, 00635089 to T.F.; 16K18358 to T.K.; 15J06369 to T.H.; 16K18583 to M.S.), MEXT KAKENHI (grant number; 15H05935 to M.Y., H.F.), Welch Foundation (grant number; I-1608 to Q.L.), NIH (grant number; GM111367 to Q.L.), Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program) from JSPS to M.Y., research grants from Uehara Memorial Foundation to M.Y., and from Takeda Science Foundation to M.Y. Nematode strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank A. Hart and H. Huang for technical advices on nematode quiescence measurement, Y. Iino for providing plasmids, M. Ikawa for providing EGxxFP plasmid, and M. Montminy and J. B. Thomas for fly stocks.
Author information
Authors and Affiliations
Contributions
H.F. and M.Y. were responsible for the overall experimental design, based on strategies conceived by M.Y. and J.S.T. M.S. constructed EEG analysis and database systems. C.M., S.K., N.H.-H., A.I., H.K., F.A., T.H., S.J.K. and K.H. conducted EEG recording and analysis. M.K. performed in situ hybridization. T.F., Se.M., F.S. and S.T. produced CRISPR-based gene-modified mice. M.A. and K.S. produced gene-modified mice. Sh.M., L.C. and Y.H. conducted roundworm experiments. T.K., H.M. and T.Y. conducted electrophysiological experiments. Z.W., J.M., A.W. and Q.L. conducted proteomics experiments. S.N., J.T. and K.K. conducted fruitfly experiments. ENU mice production and linkage analysis were conducted by I.M., T.S. and S.W. V.K. and J.S.T. designed B6 substrain-based screening. H.F. and M.Y. wrote the paper, which was reviewed by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks D.-J. Dijk, A. Sehgal and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Sleep/wakefulness screening of randomly mutagenized mice.
a, ENU-treated G0 mice were mated with B6N females to obtain the offspring. The F1 mice were used for sleep/wakefulness analysis. A mouse showing any sleep abnormalities was crossed with B6N female mice. The N2 progeny was examined for heritability of sleep abnormality and for chromosomal mapping. b, B6J (n = 20) and B6N (n = 21) showed similar total wake time (left, P = 0.67, two-tailed Student’s t-test), NREMS time (centre, P = 0.66) and REMS time (right, P = 0.84). Data are mean ± s.e.m. c, The histogram shows total daily wake time of all mice screened. Total wake time of screened mice was 735 ± 66.9 min (mean ± s.d.). Arrows indicate the founders of Sleepy mutant pedigrees.
Extended Data Figure 2 QTL analysis of Sleepy mutant pedigrees and characterization of Sik3 transcript.
a, QTL analysis of B021 (n = 119), B022 (n = 95), B024 (n = 59) and B025 (n = 112) pedigrees for total wake time produced a single LOD score peak on chromosome 9. b, Direct sequencing of the exon 12/13 boundary and exon 13/14 boundary of Sik3 mRNA of Sik13+/+ mouse. Direct sequencing of the short RT–PCR product specific to Sik3 mutant mice shows the direct transition from exon 12 to exon 14. c, d, Sik3 mRNA is expressed broadly in forebrain neurons (c). Sik3 mRNA is expressed throughout the cerebral cortex in the primary motor area (d). DG, dentate gyrus; LV, lateral ventricle; MHb, medial habenula. Scale bars, 1 mm (c) and 250 μm (d). e, RT–PCR of Sik3 mRNA from cerebral cortex and liver of Sik3+/+, Sik3Slp/+ and Sik3Slp/Slp mice. Normal Sik3 variant lacking exon 15 expressed in the cerebral cortex.
Extended Data Figure 3 Sleep/wakefulness of Sik3Slp knock-in mice.
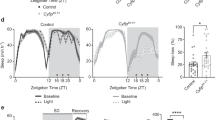
a, The structure of the Sik3 genome and targeting vector for Sik3Slp. Neomycin resistance gene under the mouse phosphoglycerol kinase promoter (neo) was sandwiched with the flippase recognition target (FRT) sequences. The guanine at the fifth nucleotide from the beginning of the intron 13 was substituted with adenine. The neo cassette was deleted by crossing with ActbCAG-FLPknock-in mice. b, RT–PCR of Sik3 mRNA of Sik3Slp/+ knock-in mice. c, Total wake time of Sik3Slp/+ knock-in mice (n = 10) and Sik3+/+ littermates (n = 6). ***P < 0.001, two-way ANOVA followed by Tukey’s test. Data are mean ± s.e.m.
Extended Data Figure 4 Sleep/wakefulness behaviours of Sik3 mutant mice.
a, Representative 8-s EEG and EMG for wake, NREMS and REMS of Sik3 mutant mice. b, Representative hypnogram of Sik3 mutant mice. Wake (blue), NREMS (green) and REMS (red) are indicated from ZT0 to ZT24. c–g, Total wake time (c), NREMS time (d), REMS time (e), NREMS/total sleep ratio (f), REMS/total sleep ratio (g) and circadian variation of REMS (h) of Sik3+/+ (n = 22), Sik3Slp/+ (n = 32) and Sik3Slp/Slp (n = 31) mice. *P < 0.05; **P < 0.01; ***P < 0.001, two-way ANOVA followed by Tukey’s test (c–g). *P < 0.05 (red); *P < 0.001 (black), one-way repeated measures ANOVA followed by Tukey’s test (h). i, Total wake time of female Sik3+/+ (n = 10), Sik3Slp/+ (n = 11) and Sik3Slp/Slp (n = 9) mice. ***P < 0.001, two-way ANOVA followed by Tukey’s test. Data are mean ± s.e.m.
Extended Data Figure 5 Characterization of sleep/wakefulness behaviours of Sik3 mutant mice.
a, Wake time after cage change at ZT15 in Sik3+/+ (n = 5), Sik3Slp/+ (n = 10) and Sik3Slp/Slp (n = 5) mice. The graph shows time spent in wakefulness from ZT15 to ZT16 under a basal condition and after cage change from the home cage to a new cage at ZT15. *P < 0.05; ***P < 0.001 versus Sik3+/+; #P < 0.05; ###P < 0.001, one-way repeated measures ANOVA followed by Tukey’s test. b, Wake time increases for 3 h after modafinil injection at ZT0 in Sik3+/+ (n = 6), Sik3Slp/+ (n = 6) and Sik3Slp/Slp (n = 6) mice. *P < 0.05; versus modafinil 10 mg kg−1 in the same genotype, #P < 0.05, ##P < 0.01, two-way ANOVA followed by Tukey’s test. c, The circadian period under constant darkness in Sik3+/+ (n = 8), Sik3Slp/+ (n = 8) and Sik3Slp/Slp (n = 6) mice. P = 0.97, one-way ANOVA. d, Total wake time of Sik3+/+ (n = 9) and Sik3Slp/+ (n = 12) mice under constant darkness. ***P < 0.001, two-tailed Student’s t-test. e, EEG power spectra of Sik3+/+ (n = 22), Sik3Slp/+ (n = 32) and Sik3Slp/Slp (n = 31) mice. *P < 0.05; ***P < 0.001, one-way ANOVA followed by Tukey’s test. f, Increase in NREMS delta power after 2 h, 4 h and 6 h of sleep deprivation of Sik3+/+ (n = 11) and Sik3Slp/+ (n = 11) mice relative to mean NREMS delta power during basal sleep. **P < 0.01, two-way ANOVA followed by Tukey’s test. g, Phosphorylation of Flag–SIK3 of Flag-Sik3+/+ brains and of Flag–SIK3(SLP) of Flag-Sik3Slp/+ brains with or without 4-h sleep deprivation. *P < 0.05; ***P < 0.001, two-way ANOVA followed by Tukey’s test. Data are mean ± s.e.m.
Extended Data Figure 6 Characterization of Flag-Sik3 mice made by CRISPR/Cas9 technology.
a, Exon 1 of the Sik3 gene contains the first and second methionine residues. The single-guide RNA was designed to target the second methionine-coding region. The donor oligonucleotide has a Flag-haemagglutinin (HA)-coding sequence immediately after the second methionine and 70-nucleotide long arms at both 5′ and 3′ ends. The Flag-HA-coding region is followed by an XbaI site. b, Immunoblotting of brain homogenates of Sik3+/+, Sik3Flag/Flag Sik3Flag,Slp/+ mice showed that anti-Flag antibody detected Flag–SIK3 protein of Sik3Flag/Flag brains and Flag–SIK3 (SLP) protein of Sik3Flag,Slp/+ brains, whereas anti-SIK3 antibody detected SIK3 proteins of all genotypes. c, RT–PCR of brain Sik3 mRNA of Sik3+/+, Sik3Flag/Flag, Sik3Flag,Slp/+ mice. d, Tryptic peptides of immunoprecipitated and gel-purified Flag–SIK3 protein were analysed by LC–MS and mapped on the reference SIK3 protein. The peptide fragments were mapped on almost entire SIK3 protein with high confidence.
Extended Data Figure 8 Identification of Nalcn mutation of the Dreamless mutant pedigree.
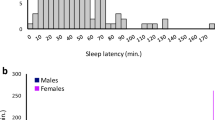
a, Histogram of REMS episode duration in N2 littermates of Dreamless mutant pedigree (bars) and all F1 mice examined (curve). b, Haplotype analysis of chromosome 14 of Dreamless mutant pedigree with or without short REMS episode duration. c, Whole-exome sequencing of Dreamless mutant N2 mice. All mice with short REMS episode duration had the single nucleotide substitution in exon 9 of the Nalcn gene.
Extended Data Figure 9 Sleep/wakefulness behaviour of Nalcn mutant mice.
a, Representative 8-s EEG and EMG for wake, NREMS and REMS of Nalcn mutant mice b, Representative hypnogram of Nalcn+/+ mice (top) and NalcnDrl/+ mice (bottom). Wake (blue), NREMS (green) and REMS (red) are indicated from ZT0 to ZT12. c, Enlarged hypnogram of around ZT7 showed the frequent transitions between NREMS and REMS of NalcnDrl/+ mice. d, Total wake time and NREMS time of NalcnDrl/+ mice (n = 29) and Nalcn+/+ mice (n = 25). Wake, P = 0.58; NREMS, P = 0.17, one-way ANOVA. e, f, Circadian period length (e) and amplitude of circadian behaviour (f) in constant darkness of NalcnDrl/+ mice (n = 6) and Nalcn+/+ mice (n = 7). P = 0.76 (e); ***P < 0.001 (f), two-tailed Student’s t-test. g, Total REMS time of NalcnDrl/+ mice (n = 9) and Nalcn+/+ mice in constant darkness (n = 8). ***P < 0.001, two-tailed Student’s t-test. h, EEG power spectra of NalcnDrl/+ mice (n = 29) and Nalcn+/+ mice (n = 25). ***P < 0.001, one-way ANOVA followed by Tukey’s test. Data are mean ± s.e.m.
Extended Data Figure 10 Increased conductance of NALCN(DRL).
a–c, Nalcn mRNA is expressed in the ventrolateral periaqueductal grey mater (vlPAG) and deep mesencephalic nucleus (DpMe) of the upper pons (a), the lateral dorsal tegmental nucleus (LDT) and sublateral dorsal nucleus (SLD) of the lower pons (b), and the lateral paragigantocellular nucleus (LPGi) of the medulla (c). AQ, aqueduct; dscp, decussation of superior cerebellar peduncle; IO, inferior olive; scp, superior cerebellar peduncle. Scale bars, 500 μm. d, Representative traces of membrane currents in response to ramp pulses (Vh = 0 mV, from −100 mV to +100 mV in 1 s; lower) recorded from HEK293T cells cotransfected with UNC80, SRC(Tyr529Phe), and NALCN–GFP (top) or NALCN(DRL)–GFP (middle). The traces are averaged from three trials. The transient capacitance currents are also recorded. e, Mean current density in response to ramp pulses (NALCN, n = 5, black line; NALCN(DRL), n = 7, purple line). The data from NALCN are also shown on an expanded scale (bottom right). f, The charge transfer of NALCN(DRL)-transfected cells was larger than that of NALCN(WT)-transfected cells. **P < 0.01, Mann–Whitney U test. The recording data are same as in e. Data are mean ± s.e.m.
Supplementary information
Supplementary Information
The file contains the raw data for Figure 1,f,h,i, and Extended Data Figures 2e, 3b, 6b,c. (PDF 1678 kb)
Rights and permissions
About this article
Cite this article
Funato, H., Miyoshi, C., Fujiyama, T. et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539, 378–383 (2016). https://doi.org/10.1038/nature20142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20142
This article is cited by
-
The collateral activity of RfxCas13d can induce lethality in a RfxCas13d knock-in mouse model
Genome Biology (2023)
-
SIK3-HDAC4 signaling pathway: the switch for transition between sleep and wakefulness
Molecular Biomedicine (2023)
-
Tfap2b acts in GABAergic neurons to control sleep in mice
Scientific Reports (2023)
-
Biochemical pathways of sleep
Cell Research (2023)
-
Noradrenergic tone is not required for neuronal activity-induced rebound sleep in zebrafish
Journal of Comparative Physiology B (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.