Abstract
Birth weight (BW) has been shown to be influenced by both fetal and maternal factors and in observational studies is reproducibly associated with future risk of adult metabolic diseases including type 2 diabetes (T2D) and cardiovascular disease1. These life-course associations have often been attributed to the impact of an adverse early life environment. Here, we performed a multi-ancestry genome-wide association study (GWAS) meta-analysis of BW in 153,781 individuals, identifying 60 loci where fetal genotype was associated with BW (P < 5 × 10−8). Overall, approximately 15% of variance in BW was captured by assays of fetal genetic variation. Using genetic association alone, we found strong inverse genetic correlations between BW and systolic blood pressure (Rg = −0.22, P = 5.5 × 10−13), T2D (Rg = −0.27, P = 1.1 × 10−6) and coronary artery disease (Rg = −0.30, P = 6.5 × 10−9). In addition, using large -cohort datasets, we demonstrated that genetic factors were the major contributor to the negative covariance between BW and future cardiometabolic risk. Pathway analyses indicated that the protein products of genes within BW-associated regions were enriched for diverse processes including insulin signalling, glucose homeostasis, glycogen biosynthesis and chromatin remodelling. There was also enrichment of associations with BW in known imprinted regions (P = 1.9 × 10−4). We demonstrate that life-course associations between early growth phenotypes and adult cardiometabolic disease are in part the result of shared genetic effects and identify some of the pathways through which these causal genetic effects are mediated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
03 October 2016
The Supplementary Information pdf was replaced and the description updated.
References
Barker, D. J. The developmental origins of chronic adult disease. Acta Paediatr. Suppl. 93, 26–33 (2004)
The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
The UK10K Project Consortium The UK10K project identifies rare variants in health and disease. Nature 526, 82–90 (2015)
Tyrrell, J. S., Yaghootkar, H., Freathy, R. M., Hattersley, A. T. & Frayling, T. M. Parental diabetes and birthweight in 236,030 individuals in the UK Biobank study. Int. J. Epidemiol. 42, 1714–1723 (2013)
Horikoshi, M. et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 45, 76–82 (2013)
Eaves, L. J., Pourcain, B. S., Smith, G. D., York, T. P. & Evans, D. M. Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”). Behav. Genet. 44, 445–455 (2014)
Feenstra, B., et al. Maternal genome-wide association study identifies a fasting glucose variant associated with offspring birth weight. Preprint at: http://biorxiv.org/content/early/2015/12/11/034207 (2015)
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015)
Segrè, A. V. et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 6, e1001058 (2010)
Baran, Y. et al. The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 25, 927–936 (2015)
Haig, D. & Westoby, M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 134, 147–155 (1989)
Peters, J. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 15, 517–530 (2014)
Johnson, T . et al. Blood pressure loci identified with a gene-centric array. Am. J. Hum. Genet. 89, 688–700 (2011)
International Consortium for Blood Pressure Genome-Wide Association Studies et al.. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478, 103–109 (2011)
Zhang, G . et al. Assessing the causal relationship of maternal height on birth size and gestational age at birth: a Mendelian randomization analysis. PLoS Med. 12, e1001865 (2015)
Tyrrell, J. et al. Genetic evidence for causal relationships between maternal obesity-related traits and birth weight. J. Am. Med. Assoc. 315, 1129–1140 (2016)
Locke, A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015)
Diver, L. A. et al. Common polymorphisms at the CYP17A1 locus associate with steroid phenotype: support for blood pressure genome-wide association study signals at this locus. Hypertension 67, 724–732 (2016)
Picado-Leonard, J. & Miller, W. L. Cloning and sequence of the human gene for P450c17 (steroid 17 alpha-hydroxylase/17,20 lyase): similarity with the gene for P450c21. DNA 6, 439–448 (1987)
Pezzi, V., Mathis, J. M., Rainey, W. E. & Carr, B. R. Profiling transcript levels for steroidogenic enzymes in fetal tissues. J. Steroid Biochem. Mol. Biol. 87, 181–189 (2003)
Escobar, J. C., Patel, S. S., Beshay, V. E., Suzuki, T. & Carr, B. R. The human placenta expresses CYP17 and generates androgens de novo. J. Clin. Endocrinol. Metab. 96, 1385–1392 (2011)
Reynolds, R. M. et al. Programming of hypertension: associations of plasma aldosterone in adult men and women with birthweight, cortisol, and blood pressure. Hypertension 53, 932–936 (2009)
CARDIoGRAMplusC4D Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 45, 25–33 (2013)
DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46, 234–244 (2014)
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014)
Hattersley, A. T. & Tooke, J. E. The fetal insulin hypothesis: an alternative explanation of the association of low birth weight with diabetes and vascular disease. Lancet 353, 1789–1792 (1999)
Dimas, A. S. et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes 63, 2158–2171 (2014)
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012)
Hattersley, A. T. et al. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat. Genet. 19, 268–270 (1998)
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010)
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 44, 955–959 (2012)
Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014)
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006)
Devlin, B. & Roeder, K. Genomic control for association studies. Biometrics 55, 997–1004 (1999)
Kang, H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010)
Allen, N. E., Sudlow, C., Peakman, T. & Collins, R. UK Biobank data: come and get it. Sci. Transl. Med. 6, 224ed4 (2014)
Loh, P. R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Mägi, R. & Morris, A. P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11, 288 (2010)
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010)
Ioannidis, J. P., Patsopoulos, N. A. & Evangelou, E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2, e841 (2007)
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375, S1–S3 (2012)
GTEx Consortium. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015)
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013)
Montgomery, S. B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010)
Schadt, E. E. et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107 (2008)
Gibbs, J. R. et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 6, e1000952 (2010)
Stranger, B. E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007)
Li, Q. et al. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum. Mol. Genet. 23, 5294–5302 (2014)
Westra, H. J. et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 45, 1238–1243 (2013)
Zou, F. et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 8, e1002707 (2012)
Hao, K. et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 8, e1003029 (2012)
Koopmann, T. T. et al. Genome-wide identification of expression quantitative trait loci (eQTLs) in human heart. PLoS One 9, e97380 (2014)
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014)
Grundberg, E. et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 7, e1001279 (2011)
Ward, L. D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012)
Flicek, P. et al. Ensembl 2014. Nucleic Acids Res. 42, D749–D755 (2014)
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009)
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010)
Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 (2015)
The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010)
Morris, A. P. Transethnic meta-analysis of genome-wide association studies. Genet. Epidemiol. 35, 809–822 (2011)
The Wellcome Trust Case Control Consortium. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012)
Wang, X. et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum. Mol. Genet. 22, 2303–2311 (2013)
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012)
Pickrell, J. K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94, 559–573 (2014)
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010)
Urbanek, M. et al. The chromosome 3q25 genomic region is associated with measures of adiposity in newborns in a multi-ethnic genome-wide association study. Hum. Mol. Genet. 22, 3583–3596 (2013)
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015)
The International HapMap Consortiumet al.. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007)
Aschard, H., Vilhjálmsson, B. J., Joshi, A. D., Price, A. L. & Kraft, P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 96, 329–339 (2015)
Wang, L., Mousavi, P. & Baranzini, S. E. iPINBPA: an integrative network-based functional module discovery tool for genome-wide association studies. Pac. Symp. Biocomput. 255–266 (2015)
Mishra, A. & Macgregor, S. VEGAS2: software for more flexible gene-based testing. Twin Res. Hum. Genet. 18, 86–91 (2015)
Lage, K. et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25, 309–316 (2007)
Whitlock, M. C. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J. Evol. Biol. 18, 1368–1373 (2005)
Hoggart, C. J. et al. Novel approach identifies SNPs in SLC2A10 and KCNK9 with evidence for parent-of-origin effect on body mass index. PLoS Genet. 10, e1004508 (2014)
Wang, S., Yu, Z., Miller, R. L., Tang, D. & Perera, F. P. Methods for detecting interactions between imprinted genes and environmental exposures using birth cohort designs with mother-offspring pairs. Hum. Hered. 71, 196–208 (2011)
Painter, J. N. et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat. Genet. 43, 51–54 (2011)
Ganesh, S. K. et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 41, 1191–1198 (2009)
Xu, X. H. et al. Two functional loci in the promoter of EPAS1 gene involved in high-altitude adaptation of Tibetans. Sci. Rep. 4, 7465 (2014)
Huerta-Sánchez, E. et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014)
Acknowledgements
Full acknowledgements and supporting grant details can be found in the Supplementary Information.
Author information
Authors and Affiliations
Consortia
Contributions
Core analyses and writing: M.H., R.N.B., F.R.D., N.M.W., M.N.K., J.F.-T., N.R.v.Z., K.J.G., A.P.M., K.K.O., J.F.F., N.J.T., J.R.P., D.M.E., M.I.M., R.M.F. Statistical analysis in individual studies: M.H., R.N.B., F.R.D., N.M.W., M.N.K., B.F., N.G., J.P.B., D.P.S., R.L.-G., T.S.A., E.K., R.R., L.-P.L., D.L.C., Y.W., E.T., C.A.W., C.T.H., J.-J.H., N.V.-T., P.K.J., E.T.H.B., I.N., N.P., A.M., E.M.v.L., R.J., V.La., M.N., J.M.M., S.E.J., P.-R.L., K.S.R., M.A.T., J.T., A.R.W., H.Y., D.M.S., I.P., K.Pan., X.W., L.C., F.G., K.E.S., M.Mu., E.V.R.A., Z.K., S.B.-G., F.S., D.T., J.W., C.M.-G., N.R.R., E.Z., G.V.D., Y.-Y.T., H.N.K., A.P.M., J.F.F., N.J.T., J.R.P., D.M.E., R.M.F. GWAS look-up in unpublished datasets: K.T.Z., N.R., D.R.N., R.C.W.M., C.H.T.T., W.H.T., S.K.G., F.J.v.R. Sample collection and data generation in individual studies: F.R.D., M.N.K., B.F., N.G., J.P.B., D.P.S., R.L.-G., R.R., L.-P.L., J.-J.H., I.N., E.M.v.L., M.B., P.M.-V., A.J.B., L.P., P.K., M.A., S.M.W., F.G., C.E.v.B., G.W., E.V.R.A., C.E.F., C.T., C.M.T., M.Sta., Z.K., D.M.H., M.V.H., H.G.d.H., F.R.R., C.M.-G., S.M.R., G.H., G.M., N.R.R., C.J.G., C.L., J.L., R.A.S., J.H.Z., F.D.M., W.L.L.Jr, A.T., M.Stu., V.Li., T.A.L., C.M.v.D., A.K., T.I.S., H.N., K.Pah., O.T.R., E.Z., G.V.D., S.-M.S., M.Me., H.C., J.F.W., M.V., J.-C.H., T.H., S.S., L.J.B., J.P.N., C.E.P., L.S.A., J.B.B., K.L.M., J.G.E., E.E.W., M.K., J.S.V., T.L., P.V., K.B., H.B., D.O.M.-K., F.R., A.G.U., C.Pi., O.P., N.J.W., H.H., V.W.J., S.F.G., A.A.V., D.A.L., G.D.S., K.K.O., J.F.F., N.J.T., J.R.P., M.I.M. Functional follow-up experiment: L.A.D., S.M.M., R.M.R., E.D., B.R.W. Individual study design and principal investigators: J.P.B., I.N., M.A., F.D.M., W.L.L.Jr, A.T., M.Stu., V.Li., T.A.L., C.M.v.D., W.K., A.K., T.I.S., H.N., K.Pah., O.T.R., G.V.D., Y.-Y.T., S.-M.S., M.Me., H.C., J.F.W., M.V., E.J.d.G., D.I.B., H.N.K., J.-C.H., T.H., A.T.H., L.J.B., J.P.N., C.E.P., J.H., L.S.A., J.B.B., K.L.M., J.G.E., E.E.W., M.K., J.S.V., T.L., P.V., K.B., H.B., D.O.M-K., A.H., F.R., A.G.U., C.Pi., O.P., C.Po., E.H., N.J.W., H.H., V.W.J., M.-R.J., S.F.G., A.A.V., T.M.F., A.P.M., K.K.O., N.J.T., J.R.P., M.I.M., R.M.F.
Corresponding authors
Ethics declarations
Competing interests
K.Z. has a scientific collaboration with Bayer HealthCare Ltd. and Population Diagnostics Inc.
Additional information
Summary statistics from the meta-analyses are available at http://egg-consortium.org/.
Reviewer Information Nature thanks J. Whitfield and the other anonymous reviewer(s) for their contribution to the peer review of this work.
A list of consortium members appears in the Supplementary Information.
A list of consortium members appears in the Supplementary Information.
Extended data figures and tables
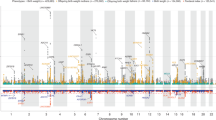
Extended Data Figure 2 Manhattan and quantile–quantile (QQ) plots of the trans-ancestry meta-analysis for BW.
a, Manhattan (main panel) and QQ (top right) plots of genome-wide association results for BW from trans-ancestry meta-analysis of up to 153,781 individuals. The association P value (on −log10 scale) for each of up to 22,434,434 SNPs (y axis) was plotted against the genomic position (NCBI Build 37; x axis). Association signals that reached genome-wide significance (P < 5 × 10−8) are shown in green if novel and pink if previously reported. In the QQ plot, the black dots represent observed P values and the grey line represents expected P values under the null distribution. The red dots represent observed P values after excluding the previously identified signals5. b, Manhattan (main panel) and QQ (top right) plots of trans-ethnic GWAS meta-analysis for BW highlighting the reported imprinted regions described in Supplementary Table 14. Novel association signals that reached genome-wide significance (P < 5 × 10−8) and mapped to imprinted regions are shown in green. Genomic regions outside imprinted regions are shaded in grey. SNPs in the imprinted regions are shown in light blue or dark blue, depending on chromosome number (odd or even). In the QQ plot, the black dots represent observed P values and the grey lines represent expected P values and their 95% confidence intervals under the null distribution for the SNPs within the imprinted regions.
Extended Data Figure 3 Regional plots for multiple distinct signals at three BW loci.
Regional plots for each locus, ZBTB7B (a), HMGA1 (b) and PTCH1 (c), are displayed from: the unconditional European-specific meta-analysis of up to 143,677 individuals (left); the approximate conditional meta-analysis for the primary signal after adjustment for the index variant for the secondary signal (middle); and the approximate conditional meta-analysis for the secondary signal after adjustment for the index variant for the primary signal (right). Directly genotyped or imputed SNPs were plotted with their association P values (on a −log10 scale) as a function of genomic position (NCBI Build 37). Estimated recombination rates (blue lines) were plotted to reflect the local linkage-disequilibrium structure around the index SNPs and their correlated proxies. SNPs were coloured in reference to linkage-disequilibrium with the particular index SNP according to a blue to red scale from R2 = 0 to 1, based on pairwise R2 values estimated from a reference of 5,000 individuals of white British origin, randomly selected from the UK Biobank.
Extended Data Figure 4 Comparison of fetal effect sizes and maternal effect sizes at 60 known and novel birth weight loci, for the first 24 loci.
The remaining loci are shown in Extended Data Fig. 5a. For each BW locus, the following six effect sizes (with 95% CI) are shown, all aligned to the same BW-raising allele: fetal_GWAS, fetal allelic effect on BW (from European ancestry meta-analysis of up to n = 143,677 individuals); fetal_unadjusted, fetal allelic effect on BW (unconditioned in n = 12,909 mother–child pairs); fetal_adjusted, fetal effect (conditioned on maternal genotype, n = 12,909); maternal_GWAS, maternal allelic effect on offspring BW (from meta-analysis of up to n = 68,254 European mothers)7; maternal_unadjusted, maternal allelic effect on offspring BW (unconditioned, n = 12,909); maternal_adjusted, maternal effect (conditioned on fetal genotype, n = 12,909). The 60 BW loci were ordered by chromosome and position (Supplementary Tables 10, 11). These plots illustrate that, in large GWAS of BW, fetal effect size estimates are larger than those of maternal at 55 out of 60 identified loci (binomial P = 1 × 10−11), suggesting that most of the associations are driven by the fetal genotype. In conditional analyses that modelled the effects of both maternal and fetal genotypes (n = 12,909 mother–child pairs), confidence intervals around the estimates were wide, precluding inference about the likely contribution of maternal versus fetal genotype at individual loci.
Extended Data Figure 5 Comparison of fetal effect sizes and maternal effect sizes at 60 known and novel birth weight loci, for the remaining 36 loci.
a, Continued from Extended Data Fig. 4. b, The scatter plot illustrates the difference between the fetal (x axis) and maternal (y axis) effect sizes in the overall maternal versus fetal GWAS results.
Extended Data Figure 6 Protein–protein Interaction (PPI) Network analysis.
a, The largest global component of BW PPI network containing 13 modules is shown. b, The histogram shows the null distribution of Z scores of BW PPI networks based on 10,000 random networks, and where the Z scores for the 13 BW modules (M1–13) lie. For each module, the two most significant GO terms are shown. c, A heat map is shown, which takes the top 50 biological processes over-represented in the global BW PPI network (listed at the right of the plot), and displays the extent of enrichment for the various trait-specific “point of contact“ (PoC) PPI networks. d, e, Trait-specific PoC PPI networks composed of proteins that are shared in both the global BW PPI network and networks generated using the same pipeline for each of the adult traits: d, canonical Wnt signalling pathway enriched for PoC PPI between BW and blood pressure (BP)-related phenotypes; and e, regulation of insulin secretion pathway enriched for PoC between BW and T2D/fasting glucose (FG). Red nodes indicate those present in PoC for BW and traits of interest; blue nodes correspond to the pathway nodes; purple nodes are those present in both the pathway and PoC; orange nodes are genes in BW loci that overlap with both the pathway and PoC. Large nodes correspond to genes in BW loci (within 300 kb from the lead SNP), and have a black border if they, amongst all BW loci, have a stronger (top 5) association with at least one of the pairing adult traits.
Extended Data Figure 7 Quantile–Quantile (QQ) plots of variance comparison between heterozygotes and homozygotes analysis in 57,715 UK Biobank samples and parent-of-origin specific analysis in 4,908 ALSPAC mother–child pairs at 59 autosomal BW loci plus DLK1.
a, QQ plot from the Quicktest analysis (ref. 77) comparing the BW variance of heterozygotes with homozygotes in 57,715 UK Biobank samples. b, QQ plot from the parent-of-origin specific analysis testing the association between BW and maternally transmitted versus paternally transmitted alleles in 4,908 mother–child pairs from the ALSPAC study (Methods, Supplementary Tables 15, 16). In both panels, the black dots represent lead SNPs at 59 identified autosomal BW loci and a further sub-genome-wide significant signal for BW near DLK1 (rs6575803; P = 5.6 × 10−8). The grey lines represent expected P values and their 95% confidence intervals under the null distribution for the 60 SNPs. Both results show trends in favour of imprinting effects at BW loci; however, despite the large sample size, these analyses were underpowered (see Methods) and much larger sample sizes are required for definitive analysis.
Extended Data Figure 8 Summary of previously reported loci for SBP, CAD, T2D and adult height and their effect on birth weight.
a–d, Effect sizes (left y axis) of previously reported 30 SBP loci13,14, 45 CAD loci23, 84 T2D loci24 and 422 adult height loci25 were plotted against effects on BW (x axis). Effect sizes were aligned to the adult trait (or risk)-raising allele. The colour of each dot indicates BW association P value: red, P < 5 × 10−8; orange, 5 × 10−8 ≤ P < 0.001; yellow, 0.001 ≤ P < 0.01; white, P ≥ 0.01. The superimposed grey frequency histogram shows the number of SNPs (right y axis) in each category of BW effect size. e, Effect sizes (with 95% CI) on BW of 45 known CAD loci were plotted arranged in the order of CAD effect size from highest to lowest, separating out the known SBP loci. CAD loci with a larger effect on BW concentrated amongst loci with primary blood pressure association. f, Effect sizes (with 95% CI) on BW of 32 known T2D loci were plotted, subdivided by previously reported categories derived from detailed adult physiological data27. Heterogeneity in BW effect sizes between five T2D loci groups with different mechanistic categories was substantial (Cochran’s Q statistic Phet = 1.2 × 10−9). In pairwise comparisons, the ‘beta cell’ group of variants differed from the other four groups: fasting hyperglycaemia (Phet = 3 × 10−11), insulin resistance (Phet = 0.002), proinsulin (Phet = 0.78) and unclassified (Phet = 0.02) groups. All of the BW effect sizes plotted in the forest plots were aligned to the trait (or risk)-raising allele.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-16 and 18-19 (see separate excel file for Supplementary Table 17), Supplementary Figures 1-2, details of grants and funding supports and a list of acknowledgements. This file was updated on 03 October 2016 to include the consortium membership lists. (PDF 2603 kb)
Supplementary Table 17
This file shows the association of BW signals with various adult metabolic and anthropometric traits. (GWAS look-ups). (XLSX 131 kb)
Supplementary Data
This file contains 60 regional plots for birth weight association. (PDF 4509 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Horikoshi, M., Beaumont, R., Day, F. et al. Genome-wide associations for birth weight and correlations with adult disease. Nature 538, 248–252 (2016). https://doi.org/10.1038/nature19806
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19806
This article is cited by
-
Maternal plasma cortisol’s effect on offspring birth weight: a Mendelian Randomisation study
BMC Pregnancy and Childbirth (2024)
-
Establishment of the early prediction models of low-birth-weight reveals influential genetic and environmental factors: a prospective cohort study
BMC Pregnancy and Childbirth (2023)
-
DNA methylation patterns at birth predict health outcomes in young adults born very low birthweight
Clinical Epigenetics (2023)
-
Exploring the novel SNPs in neuroticism and birth weight based on GWAS datasets
BMC Medical Genomics (2023)
-
Maternal prenatal cholesterol levels predict offspring weight trajectories during childhood in the Norwegian Mother, Father and Child Cohort Study
BMC Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.