Abstract
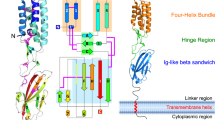
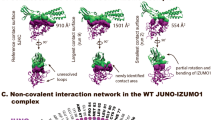
Fertilization is a fundamental process in sexual reproduction, creating a new individual through the combination of male and female gametes1,2,3,4. The IZUMO1 sperm membrane protein5 and its counterpart oocyte receptor JUNO6 have been identified as essential factors for sperm–oocyte interaction and fusion. However, the mechanism underlying their specific recognition remains poorly defined. Here, we show the crystal structures of human IZUMO1, JUNO and the IZUMO1–JUNO complex, establishing the structural basis for the IZUMO1–JUNO-mediated sperm–oocyte interaction. IZUMO1 exhibits an elongated rod-shaped structure comprised of a helical bundle IZUMO domain and an immunoglobulin-like domain that are each firmly anchored to an intervening β-hairpin region through conserved disulfide bonds. The central β-hairpin region of IZUMO1 provides the main platform for JUNO binding, while the surface located behind the putative JUNO ligand binding pocket is involved in IZUMO1 binding. Structure-based mutagenesis analysis confirms the biological importance of the IZUMO1–JUNO interaction. This structure provides a major step towards elucidating an essential phase of fertilization and it will contribute to the development of new therapeutic interventions for fertility, such as contraceptive agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The coordinates and structure-factor data of human IZUMO1, JUNO (form 1 and form 2), IZUMO1-JUNO complex (form 1, form 2, and form3) have been deposited in the Protein Data Bank under the accession numbers 5JK9, 5JKA, 5JKB, 5JKC, 5JKD and 5JKE, respectively.
References
Klinovska, K., Sebkova, N. & Dvorakova-Hortova, K. Sperm–egg fusion: a molecular enigma of mammalian reproduction. Int. J. Mol. Sci. 15, 10652–10668 (2014)
Okabe, M. The cell biology of mammalian fertilization. Development 140, 4471–4479 (2013)
Evans, J. P. Sperm–egg interaction. Annu. Rev. Physiol. 74, 477–502 (2012)
Knobil, E. & Neill, J. D. The Physiology of Reproduction. 2nd edn, (Raven Press, 1994)
Inoue, N., Ikawa, M., Isotani, A. & Okabe, M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238 (2005)
Bianchi, E., Doe, B., Goulding, D. & Wright, G. J. Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487 (2014)
Ellerman, D. A. et al. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 76, 1188–1199 (2009)
Inoue, N. et al. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development 140, 3221–3229 (2013)
Han, L. et al. Divergent evolution of vitamin B9 binding underlies Juno-mediated adhesion of mammalian gametes. Curr. Biol. 26, R100–R101 (2016)
Wibowo, A. S. et al. Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition. Proc. Natl Acad. Sci. USA 110, 15180–15188 (2013)
Chen, C. et al. Structural basis for molecular recognition of folic acid by folate receptors. Nature 500, 486–489 (2013)
Monaco, H. L. Crystal structure of chicken riboflavin-binding protein. EMBO J. 16, 1475–1483 (1997)
Aydin, H., Sultana, A., Li, S., Thavalingam, A. & Lee, J. E. molecular architecture of the human sperm Izumo1 and egg Juno fertilization complex. Nature http://dx.doi.org/10.1038/nature18595 (this issue)
Bianchi, E. & Wright, G. J. Cross-species fertilization: the hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Phil. Trans. R. Soc. B 370 (2015)
Inoue, N., Hagihara, Y., Wright, D., Suzuki, T. & Wada, I. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm–egg fusion in mice. Nat. Commun. 6, 8858 (2015)
Inoue, N., Ikawa, M. & Okabe, M. Putative sperm fusion protein IZUMO and the role of N-glycosylation. Biochem. Biophys. Res. Commun. 377, 910–914 (2008)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protocols 3, 1171–1179 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 66, 22–25 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
DeLano, W. L. The PyMOL Molecular Graphics System. http://www.pymol.org (2008)
Dundas, J. et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 (2006)
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015)
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000)
Brookes, E., Demeler, B., Rosano, C. & Rocco, M. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J. 39, 423–435 (2010)
Hashiguchi, T. et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl Acad. Sci. USA 104, 19535–19540 (2007)
Acknowledgements
We thank the beamline staff members at the Photon Factory and SPring-8 for their assistance with data collection. We thank Y. Yamada and D. Liebschner for assistance with S-SAD data collection at PF-1A. This research is supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and Development (AMED). This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (U.O., S.U., N.I. and T.S.); CREST, JST (T.S.); the Takeda Science Foundation (U.O., N.I. and T.S.); the Mochida Memorial Foundation for Medical and Pharmaceutical Research (U.O.); and the Daiichi Sankyo Foundation of Life Science (U.O.).
Author information
Authors and Affiliations
Contributions
H.I. expressed and purified recombinant proteins and performed size-exclusion chromatography and isothermal titration calorimetry experiments. H.I. and U.O. performed crystallization and structure determination. E.K and S.U performed AUC analyses. N.I. performed cellular assays. U.O and T.S. directed the research and wrote the paper with assistance from all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks K. Melcher, M. Okabe and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Sequence alignments of IZUMO1 and JUNO.
Sequence alignments of IZUMO1 (top) and JUNO (bottom) from Homo sapiens, Macaca mulatta, Equus caballus, Bos tauras or Bos mutus, Oryctolagus cuniculus and Mus musculus are shown. Secondary structure elements are displayed above the sequences. The residues of the IZUMO1–JUNO interface are indicated by yellow highlighting. The sequence identity between human and mouse IZUMO1 and JUNO are 59% and 69%, respectively, for the extracellular regions used in this study. In contrast, the interface residues exhibit less conservation (35% and 53% identity for IZUMO1 and JUNO, respectively). The N-glycosylation sites are indicated by blue Y-shaped characters. Alignments were performed using Clustal Omega software (EMBL-European Bioinformatics Institute). Residues are coloured to indicate the degree of similarity: red residues are those with the highest similarity, followed by green, blue, and black (lowest similarity).
Extended Data Figure 2 Sequence alignment of JUNO and folate receptor (FR).
Sequence alignment of JUNO from Homo sapiens, Equus caballus and Mus musculus and FRα, FRβ, and FRγ from Homo sapiens. Secondary structure elements for JUNO and FR are displayed above and below the sequences, respectively. The residues involved in the folate binding in FRs are indicated by yellow highlighting. Alignments were performed using Clustal Omega software (EMBL – European Bioinformatics Institute). Residues are coloured to indicate the degree of similarity: red residues are those with the highest similarity, followed by green, blue, and black (lowest similarity).
Extended Data Figure 3 Anomalous difference Fourier maps from S-SAD data.
The anomalous difference Fourier maps from the data collected at 2.7 Å wavelength contoured with green at the 3σ level are superposed onto the refined model of the IZUMO1–JUNO complex. The disulfide bonds are shown in stick representations and indicated by green (IZUMO1) and cyan (JUNO) labels.
Extended Data Figure 4 Superpositions of IZUMO1 and JUNO structures.
a, Superposition of the six IZUMO1 molecules (chains A–F) in the asymmetric unit. b, Superposition of the two (form 1; chains A and B) and four (form 2; chains A–D) JUNO molecules in the asymmetric unit. c, Superposition of IZUMO1 (chain A) and JUNO (chain A) structures onto the IZUMO1–JUNO complex structure (form 1, form 2, and form 3). IZUMO1 (chain A) and JUNO (chain A) are coloured grey. The IZUMO1–JUNO complexes in the form 1, form 2, and form 3 (chain A–B and chain C–D) crystals are coloured green, cyan, orange, and magenta, respectively. The r.m.s.d. values for each pair of molecules are shown schematically.
Extended Data Figure 5 Cell surface expression of IZUMO1 mutants in COS-7 cells (related to Fig. 4).
Surface localization of wild-type IZUMO1 or its mutant in COS-7 cells. IZUMO1 proteins on the cell surface (green) and nuclei (blue) were stained. Scale bars, 20 μm.
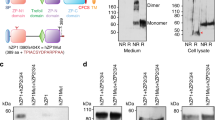
Extended Data Figure 6 Dimerization of IZUMO1 and IZUMO1–JUNO complex analysed by SV–AUC and size-exclusion chromatography.
a, b, The oligomerization states of IZUMO1 (a) and IZUMO1–JUNO complex (b) were analysed by SV–AUC at various concentrations. The normalized c(s) distributions were plotted against the sedimentation coefficients, s20,w (S). The observed sedimentation coefficient of the IZUMO1 dimer (3.2 S) at a high concentration (100 μM) in a was smaller than the expected value for IZUMO1 dimer (3.7 S), owing to the fast monomer–dimer interconversion kinetics. c, The oligomerization of IZUMO1 was analysed by gel-filtration chromatography. In each experiment, 20, 100 or 740 μM IZUMO1 (total volume of 50 μl) was injected into a Superdex 200 Increase 5/150 GL gel-filtration column (running buffer; 10 mM Tris-HCl pH 7.5 and 150 mM NaCl).
Extended Data Figure 7 Effect of reducing agent and acidic pH on the IZUMO1–JUNO interaction.
a, DTT-induced aggregation of IZUMO1. Each sample was incubated at room temperature for 2 h in the presence of 1 mM DTT, and then injected into a Superdex 200 Increase 5/150 GL gel-filtration column (running buffer; 10 mM Tris-HCl pH 7.5 and 150 mM NaCl, 1 mM DTT). Eluents were analysed by SDS–PAGE. For gel source data, see Supplementary Fig. 1. b, The interaction between IZUMO1 and JUNO at acidic pH. The IZUMO1–JUNO interaction at acidic pH was analysed by size-exclusion chromatography (SEC; top) and isothermal titration calorimetry (bottom) as in Fig. 1. In SEC analysis, each sample was injected into a Superdex 200 Increase 5/150 GL gel-filtration column (running buffer; 10 mM MES-NaOH pH 5.5 and 150 mM NaCl). Eluents were analysed by SDS–PAGE. c, The SEC chromatograms at a neutral pH of 7.5 is shown as a control experiment.
Supplementary information
Supplementary Information
This file contains the uncropped blots with size marker indications. (PDF 655 kb)
Rights and permissions
About this article
Cite this article
Ohto, U., Ishida, H., Krayukhina, E. et al. Structure of IZUMO1–JUNO reveals sperm–oocyte recognition during mammalian fertilization. Nature 534, 566–569 (2016). https://doi.org/10.1038/nature18596
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18596
This article is cited by
-
Molecular dynamics of JUNO-IZUMO1 complexation suggests biologically relevant mechanisms in fertilization
Scientific Reports (2023)
-
SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins
Communications Biology (2022)
-
Live imaging-based assay for visualising species-specific interactions in gamete adhesion molecules
Scientific Reports (2022)
-
Expression, structure and function analysis of the sperm-oocyte fusion genes Juno and Izumo1 in sheep (Ovis aries)
Journal of Animal Science and Biotechnology (2021)
-
Sperm-oocyte interplay: an overview of spermatozoon’s role in oocyte activation and current perspectives in diagnosis and fertility treatment
Cell & Bioscience (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.